
Psychrobacillus insolitus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Psychrobacillus
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
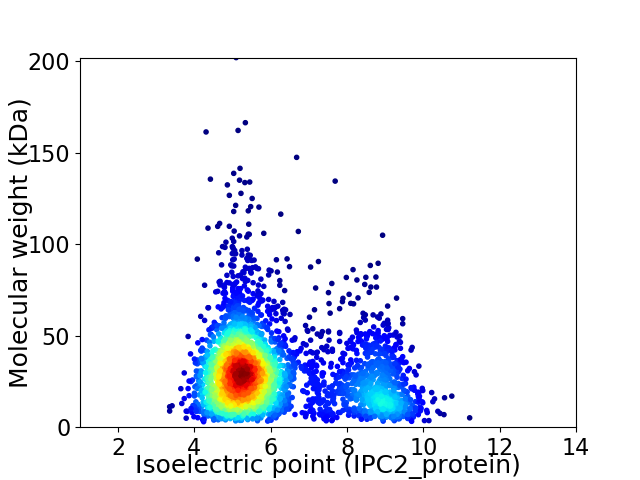
Virtual 2D-PAGE plot for 3267 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W7MGV4|A0A2W7MGV4_9BACI Uncharacterized protein OS=Psychrobacillus insolitus OX=1461 GN=C7437_10372 PE=4 SV=1
MM1 pKa = 7.34SASKK5 pKa = 10.72NSSKK9 pKa = 10.92NFLITGVLVCLFTLGGIGYY28 pKa = 8.53GFKK31 pKa = 10.72EE32 pKa = 4.2SFFAKK37 pKa = 10.49DD38 pKa = 2.94EE39 pKa = 4.91AIPVIEE45 pKa = 4.58EE46 pKa = 4.14LKK48 pKa = 9.87TLNKK52 pKa = 8.86QAPVAEE58 pKa = 4.34IEE60 pKa = 4.43TKK62 pKa = 10.38KK63 pKa = 10.87VAIQQKK69 pKa = 10.46DD70 pKa = 3.26KK71 pKa = 11.04TKK73 pKa = 10.58VIQEE77 pKa = 4.12TMPKK81 pKa = 9.81VFTILTEE88 pKa = 4.07VGQGSGFLYY97 pKa = 10.3EE98 pKa = 3.82QGGYY102 pKa = 9.7IVTNAHH108 pKa = 5.02VVAGYY113 pKa = 9.24TDD115 pKa = 3.65VIVRR119 pKa = 11.84NSAGKK124 pKa = 10.06DD125 pKa = 3.14SSAKK129 pKa = 10.5VIGISDD135 pKa = 3.73EE136 pKa = 4.32SDD138 pKa = 3.16VALLISAEE146 pKa = 4.1YY147 pKa = 10.92AKK149 pKa = 10.81VNPLEE154 pKa = 4.39TEE156 pKa = 4.07LEE158 pKa = 4.03EE159 pKa = 5.08SIVGTEE165 pKa = 4.17VIALGSPQGFEE176 pKa = 3.85NSASIGYY183 pKa = 8.03LTGVNRR189 pKa = 11.84DD190 pKa = 3.0IEE192 pKa = 4.54FGFLYY197 pKa = 10.29EE198 pKa = 4.64HH199 pKa = 7.69LYY201 pKa = 10.81QIDD204 pKa = 3.69AQIDD208 pKa = 3.73EE209 pKa = 4.88GSSGGPLIDD218 pKa = 3.89AKK220 pKa = 9.45TGKK223 pKa = 10.66VIGINSLLFKK233 pKa = 10.6EE234 pKa = 4.02NDD236 pKa = 3.24VFGFSIPMNTVTSILDD252 pKa = 3.41NWISSPMNEE261 pKa = 4.21KK262 pKa = 10.31QVTSLFGVYY271 pKa = 9.79EE272 pKa = 4.79DD273 pKa = 3.83YY274 pKa = 10.66TYY276 pKa = 11.56SDD278 pKa = 4.06EE279 pKa = 4.59NVSEE283 pKa = 3.96DD284 pKa = 3.32TYY286 pKa = 11.42YY287 pKa = 10.97EE288 pKa = 3.72EE289 pKa = 5.03DD290 pKa = 3.64AYY292 pKa = 11.26SEE294 pKa = 4.09EE295 pKa = 4.94SIPEE299 pKa = 4.07DD300 pKa = 3.47TSDD303 pKa = 4.57EE304 pKa = 3.92EE305 pKa = 4.72DD306 pKa = 3.58ANSDD310 pKa = 3.64EE311 pKa = 4.1IASYY315 pKa = 10.78EE316 pKa = 4.12EE317 pKa = 4.38EE318 pKa = 4.5SVEE321 pKa = 5.75DD322 pKa = 3.68DD323 pKa = 3.32WNDD326 pKa = 3.04NSFEE330 pKa = 4.18EE331 pKa = 4.52EE332 pKa = 4.16SLSNFIISFRR342 pKa = 11.84GYY344 pKa = 11.11YY345 pKa = 10.25EE346 pKa = 3.79MALADD351 pKa = 4.95EE352 pKa = 4.77DD353 pKa = 4.78FSWIEE358 pKa = 4.06DD359 pKa = 3.49MLGPGSIAYY368 pKa = 9.13IDD370 pKa = 3.89LEE372 pKa = 4.43KK373 pKa = 10.93YY374 pKa = 10.53VYY376 pKa = 10.11DD377 pKa = 3.55VSGQGVIFDD386 pKa = 4.22FTSNTVTAVEE396 pKa = 4.36IFDD399 pKa = 5.49DD400 pKa = 4.08YY401 pKa = 12.14ALVSTNEE408 pKa = 3.93EE409 pKa = 3.73FDD411 pKa = 3.56FTNAAGEE418 pKa = 4.33YY419 pKa = 9.94EE420 pKa = 4.49SYY422 pKa = 11.27NRR424 pKa = 11.84DD425 pKa = 2.9KK426 pKa = 10.65EE427 pKa = 4.32YY428 pKa = 10.57TVMIDD433 pKa = 3.97SEE435 pKa = 4.78GNYY438 pKa = 10.1QITDD442 pKa = 3.09IFIYY446 pKa = 10.19EE447 pKa = 3.95
MM1 pKa = 7.34SASKK5 pKa = 10.72NSSKK9 pKa = 10.92NFLITGVLVCLFTLGGIGYY28 pKa = 8.53GFKK31 pKa = 10.72EE32 pKa = 4.2SFFAKK37 pKa = 10.49DD38 pKa = 2.94EE39 pKa = 4.91AIPVIEE45 pKa = 4.58EE46 pKa = 4.14LKK48 pKa = 9.87TLNKK52 pKa = 8.86QAPVAEE58 pKa = 4.34IEE60 pKa = 4.43TKK62 pKa = 10.38KK63 pKa = 10.87VAIQQKK69 pKa = 10.46DD70 pKa = 3.26KK71 pKa = 11.04TKK73 pKa = 10.58VIQEE77 pKa = 4.12TMPKK81 pKa = 9.81VFTILTEE88 pKa = 4.07VGQGSGFLYY97 pKa = 10.3EE98 pKa = 3.82QGGYY102 pKa = 9.7IVTNAHH108 pKa = 5.02VVAGYY113 pKa = 9.24TDD115 pKa = 3.65VIVRR119 pKa = 11.84NSAGKK124 pKa = 10.06DD125 pKa = 3.14SSAKK129 pKa = 10.5VIGISDD135 pKa = 3.73EE136 pKa = 4.32SDD138 pKa = 3.16VALLISAEE146 pKa = 4.1YY147 pKa = 10.92AKK149 pKa = 10.81VNPLEE154 pKa = 4.39TEE156 pKa = 4.07LEE158 pKa = 4.03EE159 pKa = 5.08SIVGTEE165 pKa = 4.17VIALGSPQGFEE176 pKa = 3.85NSASIGYY183 pKa = 8.03LTGVNRR189 pKa = 11.84DD190 pKa = 3.0IEE192 pKa = 4.54FGFLYY197 pKa = 10.29EE198 pKa = 4.64HH199 pKa = 7.69LYY201 pKa = 10.81QIDD204 pKa = 3.69AQIDD208 pKa = 3.73EE209 pKa = 4.88GSSGGPLIDD218 pKa = 3.89AKK220 pKa = 9.45TGKK223 pKa = 10.66VIGINSLLFKK233 pKa = 10.6EE234 pKa = 4.02NDD236 pKa = 3.24VFGFSIPMNTVTSILDD252 pKa = 3.41NWISSPMNEE261 pKa = 4.21KK262 pKa = 10.31QVTSLFGVYY271 pKa = 9.79EE272 pKa = 4.79DD273 pKa = 3.83YY274 pKa = 10.66TYY276 pKa = 11.56SDD278 pKa = 4.06EE279 pKa = 4.59NVSEE283 pKa = 3.96DD284 pKa = 3.32TYY286 pKa = 11.42YY287 pKa = 10.97EE288 pKa = 3.72EE289 pKa = 5.03DD290 pKa = 3.64AYY292 pKa = 11.26SEE294 pKa = 4.09EE295 pKa = 4.94SIPEE299 pKa = 4.07DD300 pKa = 3.47TSDD303 pKa = 4.57EE304 pKa = 3.92EE305 pKa = 4.72DD306 pKa = 3.58ANSDD310 pKa = 3.64EE311 pKa = 4.1IASYY315 pKa = 10.78EE316 pKa = 4.12EE317 pKa = 4.38EE318 pKa = 4.5SVEE321 pKa = 5.75DD322 pKa = 3.68DD323 pKa = 3.32WNDD326 pKa = 3.04NSFEE330 pKa = 4.18EE331 pKa = 4.52EE332 pKa = 4.16SLSNFIISFRR342 pKa = 11.84GYY344 pKa = 11.11YY345 pKa = 10.25EE346 pKa = 3.79MALADD351 pKa = 4.95EE352 pKa = 4.77DD353 pKa = 4.78FSWIEE358 pKa = 4.06DD359 pKa = 3.49MLGPGSIAYY368 pKa = 9.13IDD370 pKa = 3.89LEE372 pKa = 4.43KK373 pKa = 10.93YY374 pKa = 10.53VYY376 pKa = 10.11DD377 pKa = 3.55VSGQGVIFDD386 pKa = 4.22FTSNTVTAVEE396 pKa = 4.36IFDD399 pKa = 5.49DD400 pKa = 4.08YY401 pKa = 12.14ALVSTNEE408 pKa = 3.93EE409 pKa = 3.73FDD411 pKa = 3.56FTNAAGEE418 pKa = 4.33YY419 pKa = 9.94EE420 pKa = 4.49SYY422 pKa = 11.27NRR424 pKa = 11.84DD425 pKa = 2.9KK426 pKa = 10.65EE427 pKa = 4.32YY428 pKa = 10.57TVMIDD433 pKa = 3.97SEE435 pKa = 4.78GNYY438 pKa = 10.1QITDD442 pKa = 3.09IFIYY446 pKa = 10.19EE447 pKa = 3.95
Molecular weight: 49.63 kDa
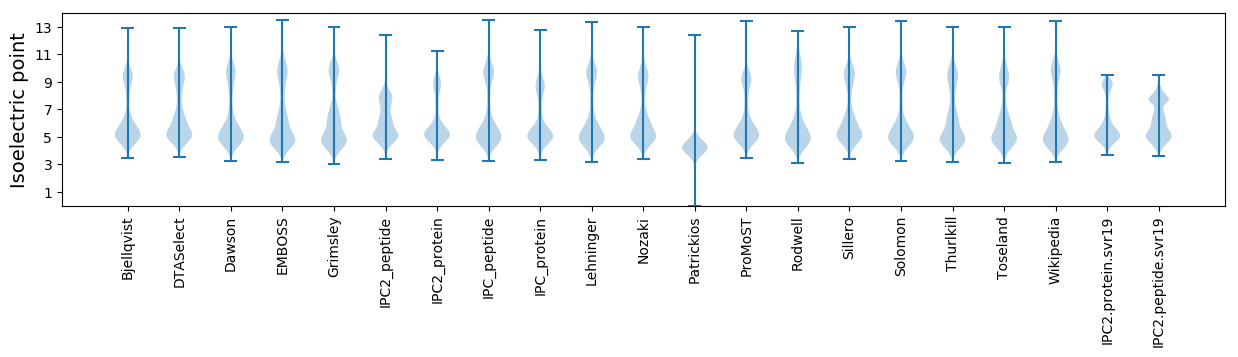
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W7N0U0|A0A2W7N0U0_9BACI Uncharacterized protein OS=Psychrobacillus insolitus OX=1461 GN=C7437_105172 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNTRR10 pKa = 11.84KK11 pKa = 9.67RR12 pKa = 11.84AKK14 pKa = 9.79NHH16 pKa = 4.64GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 9.52NGRR28 pKa = 11.84RR29 pKa = 11.84IIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNTRR10 pKa = 11.84KK11 pKa = 9.67RR12 pKa = 11.84AKK14 pKa = 9.79NHH16 pKa = 4.64GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 9.52NGRR28 pKa = 11.84RR29 pKa = 11.84IIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
919246 |
27 |
1756 |
281.4 |
31.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.008 ± 0.042 | 0.645 ± 0.013 |
5.005 ± 0.033 | 7.666 ± 0.056 |
4.591 ± 0.039 | 6.712 ± 0.039 |
1.986 ± 0.021 | 8.297 ± 0.039 |
6.809 ± 0.039 | 9.752 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.801 ± 0.022 | 4.458 ± 0.033 |
3.433 ± 0.025 | 3.699 ± 0.033 |
3.752 ± 0.033 | 6.131 ± 0.029 |
5.696 ± 0.026 | 7.236 ± 0.035 |
0.939 ± 0.015 | 3.385 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
