
Mesoplasma florum (strain ATCC 33453 / NBRC 100688 / NCTC 11704 / L1) (Acholeplasma florum)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Entomoplasmatales; Entomoplasmataceae; Mesoplasma; Mesoplasma florum
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
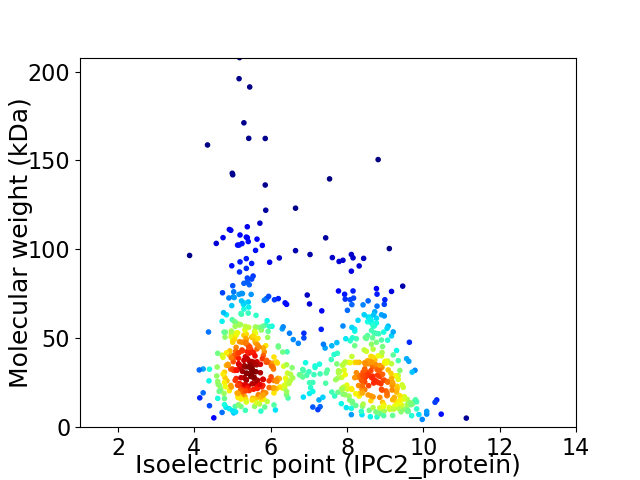
Virtual 2D-PAGE plot for 683 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q6F101|YQGF_MESFL Putative pre-16S rRNA nuclease OS=Mesoplasma florum (strain ATCC 33453 / NBRC 100688 / NCTC 11704 / L1) OX=265311 GN=Mfl464 PE=3 SV=1
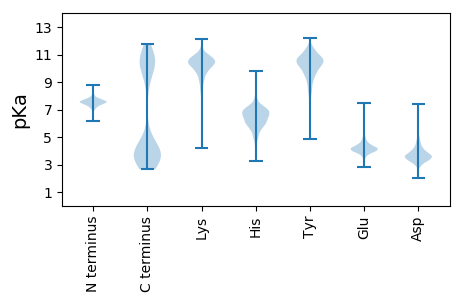
MM1 pKa = 7.56TKK3 pKa = 10.39FKK5 pKa = 11.47YY6 pKa = 10.11EE7 pKa = 3.8DD8 pKa = 2.99WLMQFINDD16 pKa = 3.53DD17 pKa = 3.38WYY19 pKa = 10.2IQINTSEE26 pKa = 3.98NNIIFDD32 pKa = 3.95EE33 pKa = 4.73VIQLHH38 pKa = 5.83EE39 pKa = 4.64KK40 pKa = 9.56WLDD43 pKa = 3.45SQDD46 pKa = 3.39YY47 pKa = 11.37NNFIKK52 pKa = 10.49EE53 pKa = 4.0NQEE56 pKa = 3.42AVAIDD61 pKa = 3.93NLPGFLEE68 pKa = 4.14NEE70 pKa = 4.52EE71 pKa = 4.16VCKK74 pKa = 9.66TDD76 pKa = 3.64EE77 pKa = 4.44YY78 pKa = 10.75IKK80 pKa = 10.98SFISGVFHH88 pKa = 7.13LRR90 pKa = 11.84IDD92 pKa = 3.58GLYY95 pKa = 10.43NIASDD100 pKa = 3.82YY101 pKa = 11.52VNAFNEE107 pKa = 4.29INEE110 pKa = 4.32HH111 pKa = 5.65SFNAVDD117 pKa = 3.59EE118 pKa = 4.81SGVDD122 pKa = 3.49VAINKK127 pKa = 9.36AFLEE131 pKa = 4.21LSEE134 pKa = 4.79KK135 pKa = 10.79YY136 pKa = 9.73YY137 pKa = 11.07EE138 pKa = 4.02EE139 pKa = 5.85LITVVRR145 pKa = 11.84NTEE148 pKa = 3.8VPDD151 pKa = 3.31EE152 pKa = 5.03FKK154 pKa = 11.26YY155 pKa = 10.58CWRR158 pKa = 11.84DD159 pKa = 3.36LIEE162 pKa = 4.13LVQRR166 pKa = 11.84FNSYY170 pKa = 8.54EE171 pKa = 4.02SRR173 pKa = 11.84EE174 pKa = 4.24DD175 pKa = 3.63KK176 pKa = 11.24LDD178 pKa = 3.39VAYY181 pKa = 10.77QLLDD185 pKa = 3.61YY186 pKa = 9.49LTTTIDD192 pKa = 3.55GFDD195 pKa = 4.48DD196 pKa = 4.72LSIDD200 pKa = 3.84LTDD203 pKa = 4.22EE204 pKa = 4.52MIEE207 pKa = 4.04SSNNFIALLIKK218 pKa = 10.1FEE220 pKa = 4.93IIFDD224 pKa = 3.85RR225 pKa = 11.84LILLKK230 pKa = 10.29EE231 pKa = 4.36HH232 pKa = 7.18IEE234 pKa = 4.04YY235 pKa = 10.59QYY237 pKa = 11.46VEE239 pKa = 4.28QKK241 pKa = 10.82GLPDD245 pKa = 3.06NFYY248 pKa = 11.31RR249 pKa = 11.84MNILDD254 pKa = 3.93RR255 pKa = 11.84YY256 pKa = 10.58KK257 pKa = 10.55EE258 pKa = 3.89IEE260 pKa = 4.04TFKK263 pKa = 10.99IMNEE267 pKa = 3.54EE268 pKa = 4.09DD269 pKa = 3.05
MM1 pKa = 7.56TKK3 pKa = 10.39FKK5 pKa = 11.47YY6 pKa = 10.11EE7 pKa = 3.8DD8 pKa = 2.99WLMQFINDD16 pKa = 3.53DD17 pKa = 3.38WYY19 pKa = 10.2IQINTSEE26 pKa = 3.98NNIIFDD32 pKa = 3.95EE33 pKa = 4.73VIQLHH38 pKa = 5.83EE39 pKa = 4.64KK40 pKa = 9.56WLDD43 pKa = 3.45SQDD46 pKa = 3.39YY47 pKa = 11.37NNFIKK52 pKa = 10.49EE53 pKa = 4.0NQEE56 pKa = 3.42AVAIDD61 pKa = 3.93NLPGFLEE68 pKa = 4.14NEE70 pKa = 4.52EE71 pKa = 4.16VCKK74 pKa = 9.66TDD76 pKa = 3.64EE77 pKa = 4.44YY78 pKa = 10.75IKK80 pKa = 10.98SFISGVFHH88 pKa = 7.13LRR90 pKa = 11.84IDD92 pKa = 3.58GLYY95 pKa = 10.43NIASDD100 pKa = 3.82YY101 pKa = 11.52VNAFNEE107 pKa = 4.29INEE110 pKa = 4.32HH111 pKa = 5.65SFNAVDD117 pKa = 3.59EE118 pKa = 4.81SGVDD122 pKa = 3.49VAINKK127 pKa = 9.36AFLEE131 pKa = 4.21LSEE134 pKa = 4.79KK135 pKa = 10.79YY136 pKa = 9.73YY137 pKa = 11.07EE138 pKa = 4.02EE139 pKa = 5.85LITVVRR145 pKa = 11.84NTEE148 pKa = 3.8VPDD151 pKa = 3.31EE152 pKa = 5.03FKK154 pKa = 11.26YY155 pKa = 10.58CWRR158 pKa = 11.84DD159 pKa = 3.36LIEE162 pKa = 4.13LVQRR166 pKa = 11.84FNSYY170 pKa = 8.54EE171 pKa = 4.02SRR173 pKa = 11.84EE174 pKa = 4.24DD175 pKa = 3.63KK176 pKa = 11.24LDD178 pKa = 3.39VAYY181 pKa = 10.77QLLDD185 pKa = 3.61YY186 pKa = 9.49LTTTIDD192 pKa = 3.55GFDD195 pKa = 4.48DD196 pKa = 4.72LSIDD200 pKa = 3.84LTDD203 pKa = 4.22EE204 pKa = 4.52MIEE207 pKa = 4.04SSNNFIALLIKK218 pKa = 10.1FEE220 pKa = 4.93IIFDD224 pKa = 3.85RR225 pKa = 11.84LILLKK230 pKa = 10.29EE231 pKa = 4.36HH232 pKa = 7.18IEE234 pKa = 4.04YY235 pKa = 10.59QYY237 pKa = 11.46VEE239 pKa = 4.28QKK241 pKa = 10.82GLPDD245 pKa = 3.06NFYY248 pKa = 11.31RR249 pKa = 11.84MNILDD254 pKa = 3.93RR255 pKa = 11.84YY256 pKa = 10.58KK257 pKa = 10.55EE258 pKa = 3.89IEE260 pKa = 4.04TFKK263 pKa = 10.99IMNEE267 pKa = 3.54EE268 pKa = 4.09DD269 pKa = 3.05
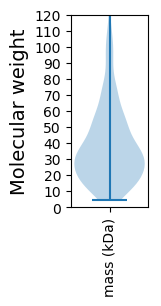
Molecular weight: 32.19 kDa
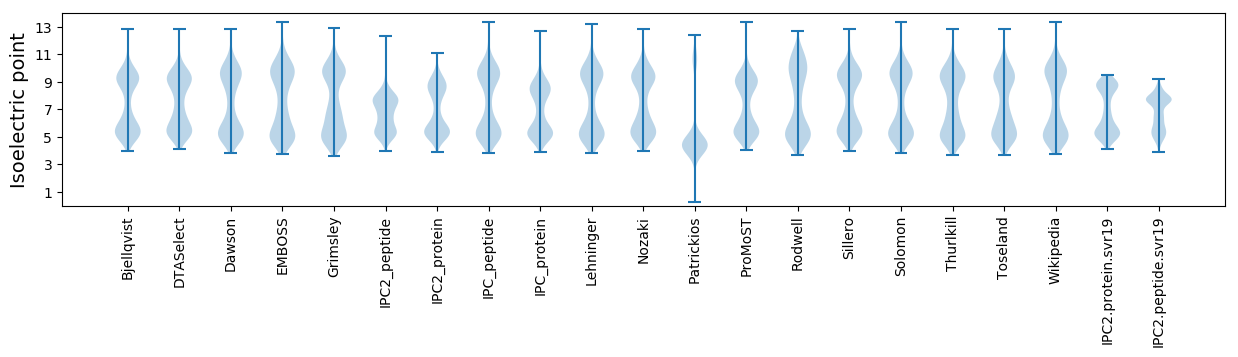
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6F0D7|Q6F0D7_MESFL Cof-like hydrolase HAD-superfamily OS=Mesoplasma florum (strain ATCC 33453 / NBRC 100688 / NCTC 11704 / L1) OX=265311 GN=Mfl680 PE=4 SV=1
MM1 pKa = 7.32KK2 pKa = 9.47RR3 pKa = 11.84TWQPSKK9 pKa = 10.02IKK11 pKa = 9.82HH12 pKa = 5.45ARR14 pKa = 11.84VHH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.41NGRR28 pKa = 11.84KK29 pKa = 9.2VIKK32 pKa = 10.14ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.33GRR39 pKa = 11.84AKK41 pKa = 9.64LTAA44 pKa = 4.21
MM1 pKa = 7.32KK2 pKa = 9.47RR3 pKa = 11.84TWQPSKK9 pKa = 10.02IKK11 pKa = 9.82HH12 pKa = 5.45ARR14 pKa = 11.84VHH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.41NGRR28 pKa = 11.84KK29 pKa = 9.2VIKK32 pKa = 10.14ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.33GRR39 pKa = 11.84AKK41 pKa = 9.64LTAA44 pKa = 4.21
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
245043 |
37 |
1814 |
358.8 |
40.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.524 ± 0.083 | 0.613 ± 0.024 |
5.355 ± 0.07 | 7.282 ± 0.1 |
5.176 ± 0.079 | 5.374 ± 0.083 |
1.243 ± 0.035 | 9.964 ± 0.086 |
9.724 ± 0.096 | 9.02 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.316 ± 0.046 | 7.252 ± 0.101 |
2.523 ± 0.044 | 3.045 ± 0.04 |
2.791 ± 0.057 | 6.52 ± 0.082 |
5.42 ± 0.079 | 6.062 ± 0.07 |
1.057 ± 0.031 | 3.739 ± 0.058 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
