
Nocardioides psychrotolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
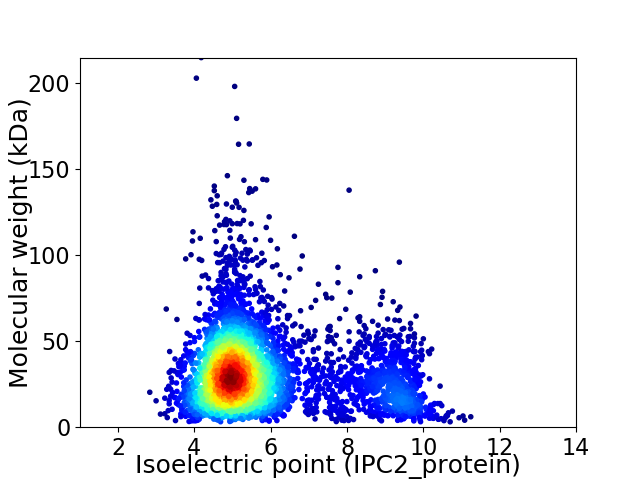
Virtual 2D-PAGE plot for 4410 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
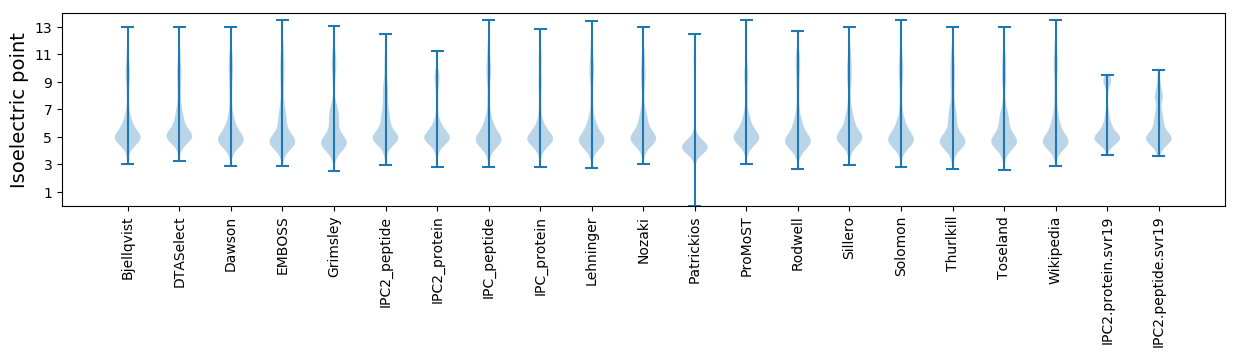
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3ETW6|A0A1I3ETW6_9ACTN Glycine cleavage system T protein (Aminomethyltransferase) OS=Nocardioides psychrotolerans OX=1005945 GN=SAMN05216561_10462 PE=3 SV=1
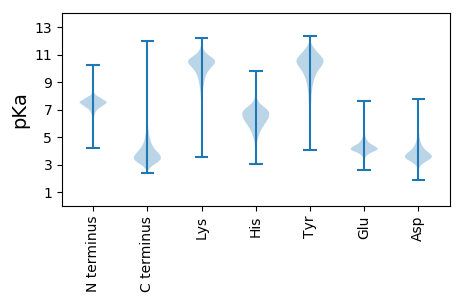
MM1 pKa = 7.88KK2 pKa = 10.1LRR4 pKa = 11.84KK5 pKa = 9.49ASAITGSLATLPLVLSACGGGDD27 pKa = 2.79IAAEE31 pKa = 4.12EE32 pKa = 4.72KK33 pKa = 10.79EE34 pKa = 4.35NEE36 pKa = 4.37SQVAAAGDD44 pKa = 3.97CGEE47 pKa = 5.26LNMAVNPWVGFAADD61 pKa = 3.98AYY63 pKa = 10.61VVGHH67 pKa = 6.62LAEE70 pKa = 4.63TEE72 pKa = 4.13LGCTVNYY79 pKa = 10.25KK80 pKa = 9.93DD81 pKa = 4.83LKK83 pKa = 11.26EE84 pKa = 4.01EE85 pKa = 4.24VSWQGFGTGEE95 pKa = 3.93VDD97 pKa = 4.81VVIEE101 pKa = 4.31DD102 pKa = 3.39WGHH105 pKa = 6.55PDD107 pKa = 3.72LEE109 pKa = 4.21KK110 pKa = 10.87LYY112 pKa = 11.08FEE114 pKa = 5.18DD115 pKa = 4.24QGGDD119 pKa = 3.28GSAVEE124 pKa = 4.62MGPTGNVGIIGWYY137 pKa = 7.4VAPWMAEE144 pKa = 4.11EE145 pKa = 4.73YY146 pKa = 10.45PDD148 pKa = 3.36ITDD151 pKa = 3.48WEE153 pKa = 4.37NLNDD157 pKa = 3.8YY158 pKa = 11.19ADD160 pKa = 3.57MFEE163 pKa = 4.36TPEE166 pKa = 4.28SGGKK170 pKa = 9.66GQFLGADD177 pKa = 3.55PSYY180 pKa = 11.13VQFDD184 pKa = 3.69EE185 pKa = 6.34AIVANLGLDD194 pKa = 3.67FQVVFSGSEE203 pKa = 3.75AASIAAFEE211 pKa = 4.09KK212 pKa = 11.09AEE214 pKa = 4.1ANKK217 pKa = 9.58TPLIGYY223 pKa = 8.72FYY225 pKa = 9.88EE226 pKa = 4.53PQWFMSEE233 pKa = 4.13VPLVKK238 pKa = 10.64VNLPEE243 pKa = 4.16FTDD246 pKa = 4.18GCQDD250 pKa = 3.2VAQDD254 pKa = 3.72VACDD258 pKa = 3.73YY259 pKa = 11.4PEE261 pKa = 4.16TQLQKK266 pKa = 10.49IVATEE271 pKa = 4.01FAEE274 pKa = 4.48SGSPAVDD281 pKa = 3.3LVSNFQWTNDD291 pKa = 3.73DD292 pKa = 3.48QNLVAKK298 pKa = 9.25YY299 pKa = 9.88ISVDD303 pKa = 3.11GMSAEE308 pKa = 4.09EE309 pKa = 4.77AAATWVEE316 pKa = 4.08EE317 pKa = 4.18NPDD320 pKa = 3.53KK321 pKa = 11.53VEE323 pKa = 3.76AWMSS327 pKa = 3.4
MM1 pKa = 7.88KK2 pKa = 10.1LRR4 pKa = 11.84KK5 pKa = 9.49ASAITGSLATLPLVLSACGGGDD27 pKa = 2.79IAAEE31 pKa = 4.12EE32 pKa = 4.72KK33 pKa = 10.79EE34 pKa = 4.35NEE36 pKa = 4.37SQVAAAGDD44 pKa = 3.97CGEE47 pKa = 5.26LNMAVNPWVGFAADD61 pKa = 3.98AYY63 pKa = 10.61VVGHH67 pKa = 6.62LAEE70 pKa = 4.63TEE72 pKa = 4.13LGCTVNYY79 pKa = 10.25KK80 pKa = 9.93DD81 pKa = 4.83LKK83 pKa = 11.26EE84 pKa = 4.01EE85 pKa = 4.24VSWQGFGTGEE95 pKa = 3.93VDD97 pKa = 4.81VVIEE101 pKa = 4.31DD102 pKa = 3.39WGHH105 pKa = 6.55PDD107 pKa = 3.72LEE109 pKa = 4.21KK110 pKa = 10.87LYY112 pKa = 11.08FEE114 pKa = 5.18DD115 pKa = 4.24QGGDD119 pKa = 3.28GSAVEE124 pKa = 4.62MGPTGNVGIIGWYY137 pKa = 7.4VAPWMAEE144 pKa = 4.11EE145 pKa = 4.73YY146 pKa = 10.45PDD148 pKa = 3.36ITDD151 pKa = 3.48WEE153 pKa = 4.37NLNDD157 pKa = 3.8YY158 pKa = 11.19ADD160 pKa = 3.57MFEE163 pKa = 4.36TPEE166 pKa = 4.28SGGKK170 pKa = 9.66GQFLGADD177 pKa = 3.55PSYY180 pKa = 11.13VQFDD184 pKa = 3.69EE185 pKa = 6.34AIVANLGLDD194 pKa = 3.67FQVVFSGSEE203 pKa = 3.75AASIAAFEE211 pKa = 4.09KK212 pKa = 11.09AEE214 pKa = 4.1ANKK217 pKa = 9.58TPLIGYY223 pKa = 8.72FYY225 pKa = 9.88EE226 pKa = 4.53PQWFMSEE233 pKa = 4.13VPLVKK238 pKa = 10.64VNLPEE243 pKa = 4.16FTDD246 pKa = 4.18GCQDD250 pKa = 3.2VAQDD254 pKa = 3.72VACDD258 pKa = 3.73YY259 pKa = 11.4PEE261 pKa = 4.16TQLQKK266 pKa = 10.49IVATEE271 pKa = 4.01FAEE274 pKa = 4.48SGSPAVDD281 pKa = 3.3LVSNFQWTNDD291 pKa = 3.73DD292 pKa = 3.48QNLVAKK298 pKa = 9.25YY299 pKa = 9.88ISVDD303 pKa = 3.11GMSAEE308 pKa = 4.09EE309 pKa = 4.77AAATWVEE316 pKa = 4.08EE317 pKa = 4.18NPDD320 pKa = 3.53KK321 pKa = 11.53VEE323 pKa = 3.76AWMSS327 pKa = 3.4
Molecular weight: 35.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3MAZ9|A0A1I3MAZ9_9ACTN Uncharacterized protein OS=Nocardioides psychrotolerans OX=1005945 GN=SAMN05216561_11534 PE=4 SV=1
MM1 pKa = 7.6GFTFNRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.12RR10 pKa = 11.84LGRR13 pKa = 11.84NTLNVSKK20 pKa = 9.51TGASLSRR27 pKa = 11.84RR28 pKa = 11.84AGPVTLSTRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84GTVRR43 pKa = 11.84LGKK46 pKa = 10.68GLGFRR51 pKa = 11.84FKK53 pKa = 11.05LL54 pKa = 3.68
MM1 pKa = 7.6GFTFNRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.12RR10 pKa = 11.84LGRR13 pKa = 11.84NTLNVSKK20 pKa = 9.51TGASLSRR27 pKa = 11.84RR28 pKa = 11.84AGPVTLSTRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84GTVRR43 pKa = 11.84LGKK46 pKa = 10.68GLGFRR51 pKa = 11.84FKK53 pKa = 11.05LL54 pKa = 3.68
Molecular weight: 6.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1380600 |
29 |
2059 |
313.1 |
33.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.719 ± 0.045 | 0.762 ± 0.01 |
6.585 ± 0.031 | 5.718 ± 0.038 |
2.837 ± 0.02 | 9.149 ± 0.035 |
2.227 ± 0.02 | 3.588 ± 0.025 |
1.959 ± 0.027 | 10.28 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.9 ± 0.014 | 1.764 ± 0.019 |
5.571 ± 0.027 | 2.847 ± 0.018 |
7.412 ± 0.038 | 5.517 ± 0.025 |
6.318 ± 0.024 | 9.433 ± 0.039 |
1.489 ± 0.016 | 1.925 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
