
Micractinium conductrix
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Trebouxiophyceae; Chlorellales; Chlorellaceae; Chlorella clade; Micractinium
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
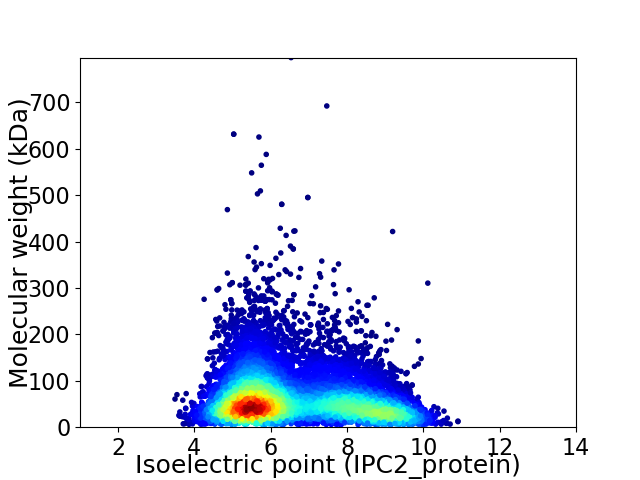
Virtual 2D-PAGE plot for 9815 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P6UZ81|A0A2P6UZ81_9CHLO Uncharacterized protein OS=Micractinium conductrix OX=554055 GN=C2E20_9200 PE=4 SV=1
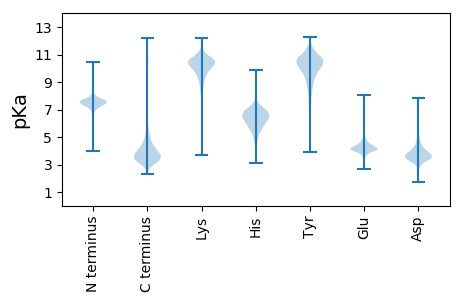
MM1 pKa = 7.53RR2 pKa = 11.84ASPAPLLVLLATLGACGGVAPARR25 pKa = 11.84VLPAEE30 pKa = 4.99DD31 pKa = 4.09PCSSLAALSPATTSAAAIVSGLGLLSGPVPAGLTLFLPSNDD72 pKa = 3.38ALASTTRR79 pKa = 11.84GGAADD84 pKa = 4.06AAQLAALSDD93 pKa = 3.68EE94 pKa = 4.57ARR96 pKa = 11.84GKK98 pKa = 9.98LAAAVLYY105 pKa = 10.88SVGAPAAGALAPDD118 pKa = 4.75ALQSAGQVTTALPDD132 pKa = 3.89APPLAVLADD141 pKa = 4.48DD142 pKa = 4.81GWFADD147 pKa = 4.0AALSLTDD154 pKa = 3.9ATNATAGVAEE164 pKa = 4.41TVQACSSVLYY174 pKa = 10.91VLDD177 pKa = 4.56KK178 pKa = 11.27VLLPAGLLDD187 pKa = 3.69PAFPSVAPDD196 pKa = 3.03QAAAILAAVPTGGTPPLSPAQPLPSPAPASGDD228 pKa = 3.63EE229 pKa = 4.31ARR231 pKa = 11.84DD232 pKa = 3.28AALALLPQLLAAQPTPGFDD251 pKa = 3.91YY252 pKa = 11.43LMLARR257 pKa = 11.84QWGPSFCAAGSCSQPAFNLFTLHH280 pKa = 7.05GLWPNNNDD288 pKa = 3.56SRR290 pKa = 11.84DD291 pKa = 3.83SPASCSGEE299 pKa = 4.25SFDD302 pKa = 5.55EE303 pKa = 4.51AALPPALLTRR313 pKa = 11.84MGCEE317 pKa = 3.58WVSLKK322 pKa = 9.54GTSASFWGYY331 pKa = 8.95EE332 pKa = 3.78WGKK335 pKa = 10.1HH336 pKa = 4.25GTCAVPGLFPNQTAYY351 pKa = 10.43FAAAMALSEE360 pKa = 3.97AWDD363 pKa = 3.54INEE366 pKa = 4.27ALAAAGLDD374 pKa = 3.62PLTARR379 pKa = 11.84SASPNDD385 pKa = 3.45VVAKK389 pKa = 10.09LQAAWGVTPLVTCSDD404 pKa = 3.36GMLQEE409 pKa = 4.08VRR411 pKa = 11.84TCFSTALQPIDD422 pKa = 3.85CPGGSGCATDD432 pKa = 4.16KK433 pKa = 10.85QQEE436 pKa = 4.2LPAGSVDD443 pKa = 3.92PPASCAQVFDD453 pKa = 4.57NVPGLQQ459 pKa = 3.21
MM1 pKa = 7.53RR2 pKa = 11.84ASPAPLLVLLATLGACGGVAPARR25 pKa = 11.84VLPAEE30 pKa = 4.99DD31 pKa = 4.09PCSSLAALSPATTSAAAIVSGLGLLSGPVPAGLTLFLPSNDD72 pKa = 3.38ALASTTRR79 pKa = 11.84GGAADD84 pKa = 4.06AAQLAALSDD93 pKa = 3.68EE94 pKa = 4.57ARR96 pKa = 11.84GKK98 pKa = 9.98LAAAVLYY105 pKa = 10.88SVGAPAAGALAPDD118 pKa = 4.75ALQSAGQVTTALPDD132 pKa = 3.89APPLAVLADD141 pKa = 4.48DD142 pKa = 4.81GWFADD147 pKa = 4.0AALSLTDD154 pKa = 3.9ATNATAGVAEE164 pKa = 4.41TVQACSSVLYY174 pKa = 10.91VLDD177 pKa = 4.56KK178 pKa = 11.27VLLPAGLLDD187 pKa = 3.69PAFPSVAPDD196 pKa = 3.03QAAAILAAVPTGGTPPLSPAQPLPSPAPASGDD228 pKa = 3.63EE229 pKa = 4.31ARR231 pKa = 11.84DD232 pKa = 3.28AALALLPQLLAAQPTPGFDD251 pKa = 3.91YY252 pKa = 11.43LMLARR257 pKa = 11.84QWGPSFCAAGSCSQPAFNLFTLHH280 pKa = 7.05GLWPNNNDD288 pKa = 3.56SRR290 pKa = 11.84DD291 pKa = 3.83SPASCSGEE299 pKa = 4.25SFDD302 pKa = 5.55EE303 pKa = 4.51AALPPALLTRR313 pKa = 11.84MGCEE317 pKa = 3.58WVSLKK322 pKa = 9.54GTSASFWGYY331 pKa = 8.95EE332 pKa = 3.78WGKK335 pKa = 10.1HH336 pKa = 4.25GTCAVPGLFPNQTAYY351 pKa = 10.43FAAAMALSEE360 pKa = 3.97AWDD363 pKa = 3.54INEE366 pKa = 4.27ALAAAGLDD374 pKa = 3.62PLTARR379 pKa = 11.84SASPNDD385 pKa = 3.45VVAKK389 pKa = 10.09LQAAWGVTPLVTCSDD404 pKa = 3.36GMLQEE409 pKa = 4.08VRR411 pKa = 11.84TCFSTALQPIDD422 pKa = 3.85CPGGSGCATDD432 pKa = 4.16KK433 pKa = 10.85QQEE436 pKa = 4.2LPAGSVDD443 pKa = 3.92PPASCAQVFDD453 pKa = 4.57NVPGLQQ459 pKa = 3.21
Molecular weight: 45.87 kDa
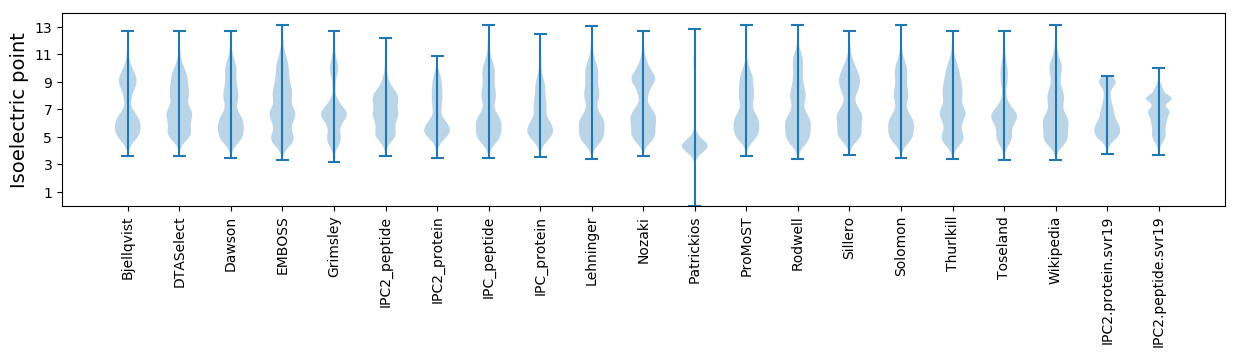
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P6VF86|A0A2P6VF86_9CHLO Nucleotide-diphospho-sugar transferase OS=Micractinium conductrix OX=554055 GN=C2E20_4110 PE=4 SV=1
MM1 pKa = 7.45PSAWQRR7 pKa = 11.84WRR9 pKa = 11.84PWLAAAANLQMRR21 pKa = 11.84AGRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84PGRR29 pKa = 11.84QRR31 pKa = 11.84LNQGHH36 pKa = 5.66QARR39 pKa = 11.84KK40 pKa = 8.97AAGLAGAEE48 pKa = 3.85AAAWATALLNARR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84AGSDD66 pKa = 3.25VKK68 pKa = 10.24WRR70 pKa = 11.84RR71 pKa = 11.84TRR73 pKa = 11.84RR74 pKa = 11.84ASARR78 pKa = 11.84RR79 pKa = 11.84SSPPGPWHH87 pKa = 7.04ANDD90 pKa = 3.35HH91 pKa = 5.99AARR94 pKa = 11.84PAVRR98 pKa = 11.84LRR100 pKa = 11.84RR101 pKa = 11.84AAAGHH106 pKa = 4.93QRR108 pKa = 3.32
MM1 pKa = 7.45PSAWQRR7 pKa = 11.84WRR9 pKa = 11.84PWLAAAANLQMRR21 pKa = 11.84AGRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84PGRR29 pKa = 11.84QRR31 pKa = 11.84LNQGHH36 pKa = 5.66QARR39 pKa = 11.84KK40 pKa = 8.97AAGLAGAEE48 pKa = 3.85AAAWATALLNARR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84AGSDD66 pKa = 3.25VKK68 pKa = 10.24WRR70 pKa = 11.84RR71 pKa = 11.84TRR73 pKa = 11.84RR74 pKa = 11.84ASARR78 pKa = 11.84RR79 pKa = 11.84SSPPGPWHH87 pKa = 7.04ANDD90 pKa = 3.35HH91 pKa = 5.99AARR94 pKa = 11.84PAVRR98 pKa = 11.84LRR100 pKa = 11.84RR101 pKa = 11.84AAAGHH106 pKa = 4.93QRR108 pKa = 3.32
Molecular weight: 12.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6408442 |
54 |
7986 |
652.9 |
69.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
16.178 ± 0.054 | 1.62 ± 0.014 |
4.451 ± 0.015 | 5.863 ± 0.027 |
2.606 ± 0.013 | 9.29 ± 0.026 |
2.148 ± 0.009 | 2.303 ± 0.014 |
3.041 ± 0.018 | 9.956 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.869 ± 0.009 | 1.898 ± 0.011 |
6.572 ± 0.026 | 5.35 ± 0.029 |
6.596 ± 0.019 | 6.397 ± 0.019 |
4.376 ± 0.016 | 6.258 ± 0.015 |
1.375 ± 0.008 | 1.854 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
