
Soft spider associated circular virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Spider associated cyclovirus 1
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
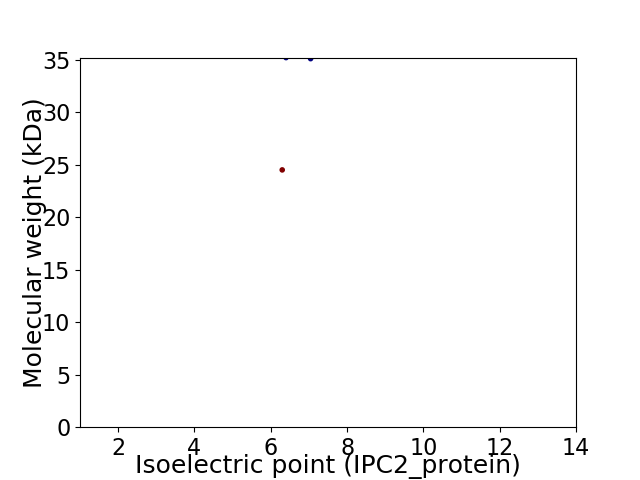
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BP97|A0A346BP97_9CIRC Putative capsid protein OS=Soft spider associated circular virus 1 OX=2293301 PE=4 SV=1
MM1 pKa = 6.28TTWPASASTADD12 pKa = 3.69AAPPDD17 pKa = 4.05GSAPSAEE24 pKa = 4.22PGEE27 pKa = 4.53AEE29 pKa = 5.29GDD31 pKa = 3.49QTDD34 pKa = 3.42AKK36 pKa = 10.12LTFTASVWLPEE47 pKa = 4.2LQSSHH52 pKa = 7.43LLVQSSATDD61 pKa = 2.78ICLRR65 pKa = 11.84VDD67 pKa = 4.56DD68 pKa = 5.54LPFNNTQIGYY78 pKa = 9.27LYY80 pKa = 10.87SHH82 pKa = 6.87LRR84 pKa = 11.84INKK87 pKa = 8.79CVVKK91 pKa = 10.34IYY93 pKa = 10.05PCGNFAEE100 pKa = 4.41QPVAGSYY107 pKa = 10.07IEE109 pKa = 4.23PEE111 pKa = 4.49VYY113 pKa = 10.56SSITYY118 pKa = 10.01DD119 pKa = 3.58PQVVDD124 pKa = 3.46ATYY127 pKa = 10.27TLTNGPTVKK136 pKa = 10.24SHH138 pKa = 6.18GPRR141 pKa = 11.84SKK143 pKa = 9.01ITRR146 pKa = 11.84VVYY149 pKa = 9.44PKK151 pKa = 10.09VCYY154 pKa = 9.18PVWGVNGDD162 pKa = 3.34ISSTSSSIRR171 pKa = 11.84NMKK174 pKa = 8.6KK175 pKa = 10.48APWMSINSAGITDD188 pKa = 3.76ACLGKK193 pKa = 10.48LRR195 pKa = 11.84IYY197 pKa = 10.14MGANSGDD204 pKa = 4.2GITKK208 pKa = 7.96VWKK211 pKa = 9.1WNVTKK216 pKa = 10.02TYY218 pKa = 10.63YY219 pKa = 10.46FQLKK223 pKa = 9.5NVV225 pKa = 3.62
MM1 pKa = 6.28TTWPASASTADD12 pKa = 3.69AAPPDD17 pKa = 4.05GSAPSAEE24 pKa = 4.22PGEE27 pKa = 4.53AEE29 pKa = 5.29GDD31 pKa = 3.49QTDD34 pKa = 3.42AKK36 pKa = 10.12LTFTASVWLPEE47 pKa = 4.2LQSSHH52 pKa = 7.43LLVQSSATDD61 pKa = 2.78ICLRR65 pKa = 11.84VDD67 pKa = 4.56DD68 pKa = 5.54LPFNNTQIGYY78 pKa = 9.27LYY80 pKa = 10.87SHH82 pKa = 6.87LRR84 pKa = 11.84INKK87 pKa = 8.79CVVKK91 pKa = 10.34IYY93 pKa = 10.05PCGNFAEE100 pKa = 4.41QPVAGSYY107 pKa = 10.07IEE109 pKa = 4.23PEE111 pKa = 4.49VYY113 pKa = 10.56SSITYY118 pKa = 10.01DD119 pKa = 3.58PQVVDD124 pKa = 3.46ATYY127 pKa = 10.27TLTNGPTVKK136 pKa = 10.24SHH138 pKa = 6.18GPRR141 pKa = 11.84SKK143 pKa = 9.01ITRR146 pKa = 11.84VVYY149 pKa = 9.44PKK151 pKa = 10.09VCYY154 pKa = 9.18PVWGVNGDD162 pKa = 3.34ISSTSSSIRR171 pKa = 11.84NMKK174 pKa = 8.6KK175 pKa = 10.48APWMSINSAGITDD188 pKa = 3.76ACLGKK193 pKa = 10.48LRR195 pKa = 11.84IYY197 pKa = 10.14MGANSGDD204 pKa = 4.2GITKK208 pKa = 7.96VWKK211 pKa = 9.1WNVTKK216 pKa = 10.02TYY218 pKa = 10.63YY219 pKa = 10.46FQLKK223 pKa = 9.5NVV225 pKa = 3.62
Molecular weight: 24.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BP97|A0A346BP97_9CIRC Putative capsid protein OS=Soft spider associated circular virus 1 OX=2293301 PE=4 SV=1
MM1 pKa = 7.32AQGKK5 pKa = 8.81RR6 pKa = 11.84WCFTLNNYY14 pKa = 8.6TDD16 pKa = 3.82GDD18 pKa = 3.95EE19 pKa = 4.06EE20 pKa = 4.41RR21 pKa = 11.84LRR23 pKa = 11.84GLDD26 pKa = 3.37VKK28 pKa = 9.0YY29 pKa = 10.51LCVGRR34 pKa = 11.84EE35 pKa = 3.88VAPTTGTRR43 pKa = 11.84HH44 pKa = 5.15LQGFVNFRR52 pKa = 11.84TCQRR56 pKa = 11.84FNKK59 pKa = 9.49ARR61 pKa = 11.84EE62 pKa = 4.12LIGTSAHH69 pKa = 6.64LEE71 pKa = 4.08LAKK74 pKa = 10.28GTDD77 pKa = 3.69VQNKK81 pKa = 8.25EE82 pKa = 4.39YY83 pKa = 9.98CCKK86 pKa = 10.48SADD89 pKa = 3.62FFEE92 pKa = 5.12VCVQGQRR99 pKa = 11.84TDD101 pKa = 3.55LAAVAAACAVRR112 pKa = 11.84GNSLEE117 pKa = 4.14KK118 pKa = 10.68VALEE122 pKa = 4.31HH123 pKa = 5.77PQEE126 pKa = 4.36FIKK129 pKa = 9.74YY130 pKa = 8.0HH131 pKa = 6.49RR132 pKa = 11.84GIQRR136 pKa = 11.84YY137 pKa = 7.98YY138 pKa = 10.59EE139 pKa = 3.79IANPIQHH146 pKa = 7.2RR147 pKa = 11.84DD148 pKa = 3.11FKK150 pKa = 10.64TRR152 pKa = 11.84VHH154 pKa = 6.01VFVGPPGSGKK164 pKa = 10.22SRR166 pKa = 11.84LANACGEE173 pKa = 4.48VNGEE177 pKa = 3.7IYY179 pKa = 10.68YY180 pKa = 10.31KK181 pKa = 10.54PRR183 pKa = 11.84GEE185 pKa = 3.68WWDD188 pKa = 3.77GYY190 pKa = 10.64RR191 pKa = 11.84DD192 pKa = 3.45QSTIVVDD199 pKa = 5.96DD200 pKa = 4.16FYY202 pKa = 11.95GWLKK206 pKa = 10.72YY207 pKa = 10.67DD208 pKa = 4.31EE209 pKa = 4.82LLKK212 pKa = 10.7ICDD215 pKa = 3.62RR216 pKa = 11.84YY217 pKa = 9.79PYY219 pKa = 9.79KK220 pKa = 11.1VPVKK224 pKa = 9.61GAYY227 pKa = 9.74VEE229 pKa = 4.65FTARR233 pKa = 11.84NIFITSNEE241 pKa = 4.73PIDD244 pKa = 3.35NWYY247 pKa = 10.1KK248 pKa = 10.75FNGYY252 pKa = 10.07DD253 pKa = 3.14PAAIKK258 pKa = 10.51RR259 pKa = 11.84RR260 pKa = 11.84VSVYY264 pKa = 9.6MFINGLEE271 pKa = 3.89PVYY274 pKa = 10.91YY275 pKa = 10.03IGPDD279 pKa = 3.96DD280 pKa = 3.79EE281 pKa = 4.37WLSLKK286 pKa = 10.51VKK288 pKa = 10.87AEE290 pKa = 4.0FKK292 pKa = 10.28IACQNVLLDD301 pKa = 4.51LDD303 pKa = 4.25NNN305 pKa = 4.12
MM1 pKa = 7.32AQGKK5 pKa = 8.81RR6 pKa = 11.84WCFTLNNYY14 pKa = 8.6TDD16 pKa = 3.82GDD18 pKa = 3.95EE19 pKa = 4.06EE20 pKa = 4.41RR21 pKa = 11.84LRR23 pKa = 11.84GLDD26 pKa = 3.37VKK28 pKa = 9.0YY29 pKa = 10.51LCVGRR34 pKa = 11.84EE35 pKa = 3.88VAPTTGTRR43 pKa = 11.84HH44 pKa = 5.15LQGFVNFRR52 pKa = 11.84TCQRR56 pKa = 11.84FNKK59 pKa = 9.49ARR61 pKa = 11.84EE62 pKa = 4.12LIGTSAHH69 pKa = 6.64LEE71 pKa = 4.08LAKK74 pKa = 10.28GTDD77 pKa = 3.69VQNKK81 pKa = 8.25EE82 pKa = 4.39YY83 pKa = 9.98CCKK86 pKa = 10.48SADD89 pKa = 3.62FFEE92 pKa = 5.12VCVQGQRR99 pKa = 11.84TDD101 pKa = 3.55LAAVAAACAVRR112 pKa = 11.84GNSLEE117 pKa = 4.14KK118 pKa = 10.68VALEE122 pKa = 4.31HH123 pKa = 5.77PQEE126 pKa = 4.36FIKK129 pKa = 9.74YY130 pKa = 8.0HH131 pKa = 6.49RR132 pKa = 11.84GIQRR136 pKa = 11.84YY137 pKa = 7.98YY138 pKa = 10.59EE139 pKa = 3.79IANPIQHH146 pKa = 7.2RR147 pKa = 11.84DD148 pKa = 3.11FKK150 pKa = 10.64TRR152 pKa = 11.84VHH154 pKa = 6.01VFVGPPGSGKK164 pKa = 10.22SRR166 pKa = 11.84LANACGEE173 pKa = 4.48VNGEE177 pKa = 3.7IYY179 pKa = 10.68YY180 pKa = 10.31KK181 pKa = 10.54PRR183 pKa = 11.84GEE185 pKa = 3.68WWDD188 pKa = 3.77GYY190 pKa = 10.64RR191 pKa = 11.84DD192 pKa = 3.45QSTIVVDD199 pKa = 5.96DD200 pKa = 4.16FYY202 pKa = 11.95GWLKK206 pKa = 10.72YY207 pKa = 10.67DD208 pKa = 4.31EE209 pKa = 4.82LLKK212 pKa = 10.7ICDD215 pKa = 3.62RR216 pKa = 11.84YY217 pKa = 9.79PYY219 pKa = 9.79KK220 pKa = 11.1VPVKK224 pKa = 9.61GAYY227 pKa = 9.74VEE229 pKa = 4.65FTARR233 pKa = 11.84NIFITSNEE241 pKa = 4.73PIDD244 pKa = 3.35NWYY247 pKa = 10.1KK248 pKa = 10.75FNGYY252 pKa = 10.07DD253 pKa = 3.14PAAIKK258 pKa = 10.51RR259 pKa = 11.84RR260 pKa = 11.84VSVYY264 pKa = 9.6MFINGLEE271 pKa = 3.89PVYY274 pKa = 10.91YY275 pKa = 10.03IGPDD279 pKa = 3.96DD280 pKa = 3.79EE281 pKa = 4.37WLSLKK286 pKa = 10.51VKK288 pKa = 10.87AEE290 pKa = 4.0FKK292 pKa = 10.28IACQNVLLDD301 pKa = 4.51LDD303 pKa = 4.25NNN305 pKa = 4.12
Molecular weight: 35.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
530 |
225 |
305 |
265.0 |
29.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.547 ± 0.27 | 2.83 ± 0.363 |
5.849 ± 0.308 | 5.094 ± 1.185 |
3.585 ± 1.079 | 7.17 ± 0.301 |
1.698 ± 0.218 | 5.283 ± 0.296 |
6.226 ± 0.268 | 6.415 ± 0.381 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.132 ± 0.386 | 5.283 ± 0.235 |
5.283 ± 1.092 | 3.396 ± 0.17 |
5.094 ± 1.45 | 6.038 ± 2.5 |
6.038 ± 1.438 | 7.925 ± 0.045 |
2.264 ± 0.24 | 5.849 ± 0.308 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
