
Anaerobacillus alkaliphilus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Anaerobacillus
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
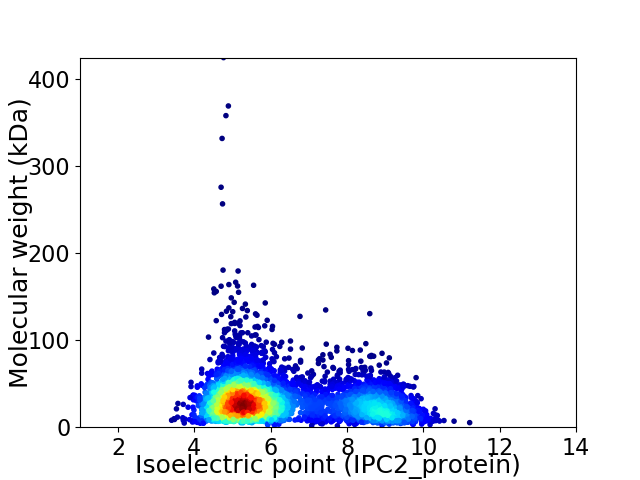
Virtual 2D-PAGE plot for 4643 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q0VRJ2|A0A4Q0VRJ2_9BACI TlyA family RNA methyltransferase OS=Anaerobacillus alkaliphilus OX=1548597 GN=DS745_15010 PE=4 SV=1
MM1 pKa = 7.47TLLKK5 pKa = 10.41NINVNIMLAVVGIFAVTVFLFMLQSHH31 pKa = 6.34SVPVDD36 pKa = 3.44QVSEE40 pKa = 4.14EE41 pKa = 4.24VTVEE45 pKa = 4.01EE46 pKa = 4.84NGNEE50 pKa = 3.86ASKK53 pKa = 11.12SEE55 pKa = 4.04NGNHH59 pKa = 7.39DD60 pKa = 3.6EE61 pKa = 4.97TEE63 pKa = 4.3TNTNDD68 pKa = 3.33NGSEE72 pKa = 4.14EE73 pKa = 4.25NEE75 pKa = 4.55SNGSDD80 pKa = 3.03IEE82 pKa = 4.24EE83 pKa = 4.37ANEE86 pKa = 3.77NTNGQEE92 pKa = 4.2YY93 pKa = 10.4LWGLDD98 pKa = 3.09SADD101 pKa = 3.85RR102 pKa = 11.84VDD104 pKa = 4.48QSFYY108 pKa = 11.47DD109 pKa = 3.7CFVTNFGEE117 pKa = 4.3PVAFGRR123 pKa = 11.84YY124 pKa = 9.29LEE126 pKa = 4.54TKK128 pKa = 10.46DD129 pKa = 3.43GVSYY133 pKa = 10.86GLSEE137 pKa = 4.41EE138 pKa = 4.25EE139 pKa = 4.66VAFLQDD145 pKa = 3.5KK146 pKa = 9.77GIKK149 pKa = 9.53IIPIYY154 pKa = 10.8NHH156 pKa = 6.92FIDD159 pKa = 3.75GTTYY163 pKa = 10.5EE164 pKa = 4.6KK165 pKa = 10.79GVQEE169 pKa = 4.79AEE171 pKa = 4.01TAITYY176 pKa = 10.69AEE178 pKa = 4.68AIGVTEE184 pKa = 4.12GVYY187 pKa = 10.15IFANVEE193 pKa = 3.62PEE195 pKa = 4.05YY196 pKa = 10.59PIDD199 pKa = 4.72ADD201 pKa = 4.87FLLGWFEE208 pKa = 4.92TIQSSQYY215 pKa = 9.83KK216 pKa = 8.71PGVYY220 pKa = 10.25GIFGEE225 pKa = 4.59GRR227 pKa = 11.84EE228 pKa = 4.03LTAAFEE234 pKa = 4.18RR235 pKa = 11.84ALGNNEE241 pKa = 4.33AIGEE245 pKa = 4.15EE246 pKa = 4.18MAVWHH251 pKa = 6.38SSEE254 pKa = 4.28PYY256 pKa = 9.92IGITTKK262 pKa = 10.23EE263 pKa = 4.22NKK265 pKa = 9.81PEE267 pKa = 3.99FAPDD271 pKa = 3.12VPDD274 pKa = 4.37FVNVSIWQYY283 pKa = 11.28GIDD286 pKa = 5.05AEE288 pKa = 4.58TCNIDD293 pKa = 3.43TNLINSEE300 pKa = 4.11IMDD303 pKa = 4.39SLWW306 pKa = 4.04
MM1 pKa = 7.47TLLKK5 pKa = 10.41NINVNIMLAVVGIFAVTVFLFMLQSHH31 pKa = 6.34SVPVDD36 pKa = 3.44QVSEE40 pKa = 4.14EE41 pKa = 4.24VTVEE45 pKa = 4.01EE46 pKa = 4.84NGNEE50 pKa = 3.86ASKK53 pKa = 11.12SEE55 pKa = 4.04NGNHH59 pKa = 7.39DD60 pKa = 3.6EE61 pKa = 4.97TEE63 pKa = 4.3TNTNDD68 pKa = 3.33NGSEE72 pKa = 4.14EE73 pKa = 4.25NEE75 pKa = 4.55SNGSDD80 pKa = 3.03IEE82 pKa = 4.24EE83 pKa = 4.37ANEE86 pKa = 3.77NTNGQEE92 pKa = 4.2YY93 pKa = 10.4LWGLDD98 pKa = 3.09SADD101 pKa = 3.85RR102 pKa = 11.84VDD104 pKa = 4.48QSFYY108 pKa = 11.47DD109 pKa = 3.7CFVTNFGEE117 pKa = 4.3PVAFGRR123 pKa = 11.84YY124 pKa = 9.29LEE126 pKa = 4.54TKK128 pKa = 10.46DD129 pKa = 3.43GVSYY133 pKa = 10.86GLSEE137 pKa = 4.41EE138 pKa = 4.25EE139 pKa = 4.66VAFLQDD145 pKa = 3.5KK146 pKa = 9.77GIKK149 pKa = 9.53IIPIYY154 pKa = 10.8NHH156 pKa = 6.92FIDD159 pKa = 3.75GTTYY163 pKa = 10.5EE164 pKa = 4.6KK165 pKa = 10.79GVQEE169 pKa = 4.79AEE171 pKa = 4.01TAITYY176 pKa = 10.69AEE178 pKa = 4.68AIGVTEE184 pKa = 4.12GVYY187 pKa = 10.15IFANVEE193 pKa = 3.62PEE195 pKa = 4.05YY196 pKa = 10.59PIDD199 pKa = 4.72ADD201 pKa = 4.87FLLGWFEE208 pKa = 4.92TIQSSQYY215 pKa = 9.83KK216 pKa = 8.71PGVYY220 pKa = 10.25GIFGEE225 pKa = 4.59GRR227 pKa = 11.84EE228 pKa = 4.03LTAAFEE234 pKa = 4.18RR235 pKa = 11.84ALGNNEE241 pKa = 4.33AIGEE245 pKa = 4.15EE246 pKa = 4.18MAVWHH251 pKa = 6.38SSEE254 pKa = 4.28PYY256 pKa = 9.92IGITTKK262 pKa = 10.23EE263 pKa = 4.22NKK265 pKa = 9.81PEE267 pKa = 3.99FAPDD271 pKa = 3.12VPDD274 pKa = 4.37FVNVSIWQYY283 pKa = 11.28GIDD286 pKa = 5.05AEE288 pKa = 4.58TCNIDD293 pKa = 3.43TNLINSEE300 pKa = 4.11IMDD303 pKa = 4.39SLWW306 pKa = 4.04
Molecular weight: 34.02 kDa
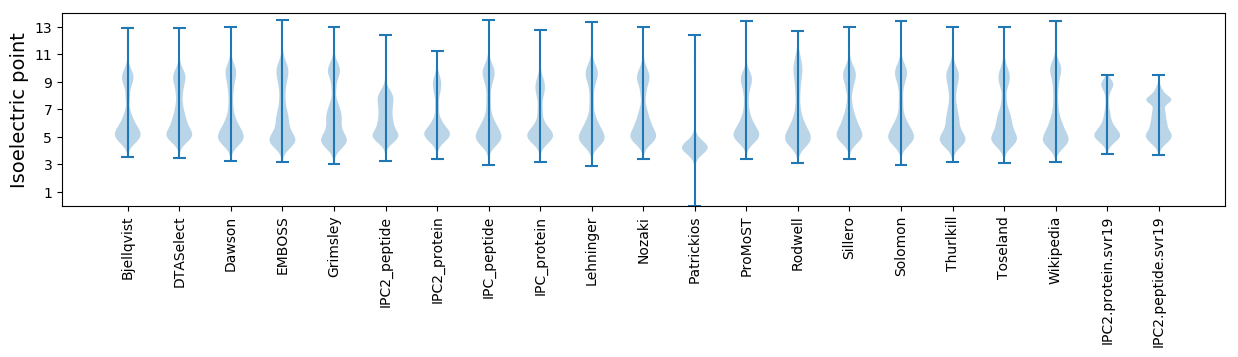
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q0VVH2|A0A4Q0VVH2_9BACI Uncharacterized protein OS=Anaerobacillus alkaliphilus OX=1548597 GN=DS745_06685 PE=4 SV=1
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.45GFRR20 pKa = 11.84ARR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.91VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.45GFRR20 pKa = 11.84ARR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.91VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1361674 |
15 |
3880 |
293.3 |
33.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.587 ± 0.046 | 0.716 ± 0.01 |
4.939 ± 0.024 | 7.554 ± 0.044 |
4.772 ± 0.028 | 6.729 ± 0.035 |
2.095 ± 0.016 | 7.993 ± 0.038 |
6.592 ± 0.034 | 9.994 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.6 ± 0.019 | 4.596 ± 0.033 |
3.54 ± 0.021 | 3.755 ± 0.026 |
4.031 ± 0.025 | 5.907 ± 0.029 |
5.638 ± 0.029 | 7.334 ± 0.035 |
1.027 ± 0.014 | 3.602 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
