
Tortoise microvirus 26
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
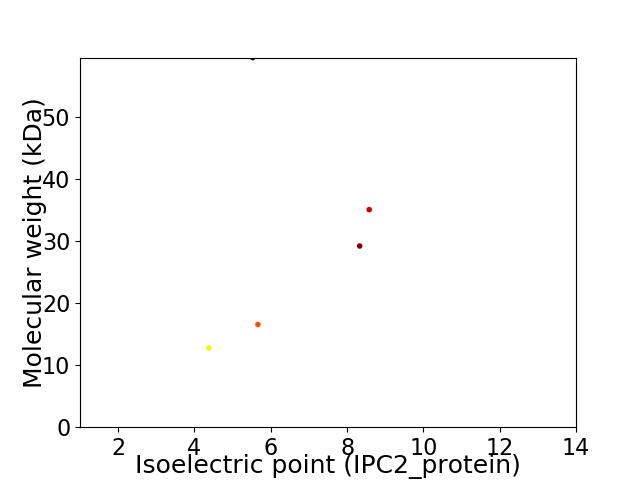
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W708|A0A4P8W708_9VIRU DNA pilot protein OS=Tortoise microvirus 26 OX=2583128 PE=4 SV=1
MM1 pKa = 7.62NKK3 pKa = 9.92RR4 pKa = 11.84IYY6 pKa = 10.45VIYY9 pKa = 9.16NTDD12 pKa = 3.37EE13 pKa = 4.19KK14 pKa = 11.26DD15 pKa = 3.71SNVFSARR22 pKa = 11.84STDD25 pKa = 3.37EE26 pKa = 4.14AFDD29 pKa = 3.66IAFQTIFDD37 pKa = 4.06TEE39 pKa = 4.12EE40 pKa = 3.6FDD42 pKa = 5.63EE43 pKa = 4.77FFNDD47 pKa = 3.19NGDD50 pKa = 3.66IVVTKK55 pKa = 10.61ANKK58 pKa = 10.2AYY60 pKa = 10.36FFEE63 pKa = 4.81CLKK66 pKa = 9.81GWKK69 pKa = 9.13MYY71 pKa = 10.73DD72 pKa = 3.22YY73 pKa = 11.45GKK75 pKa = 10.78YY76 pKa = 10.58DD77 pKa = 3.16SDD79 pKa = 3.68KK80 pKa = 11.21VLFSLTEE87 pKa = 3.85HH88 pKa = 7.16PNPIDD93 pKa = 3.38FNEE96 pKa = 4.03YY97 pKa = 10.86LKK99 pKa = 10.52ILQDD103 pKa = 3.58KK104 pKa = 10.66VVGLL108 pKa = 4.05
MM1 pKa = 7.62NKK3 pKa = 9.92RR4 pKa = 11.84IYY6 pKa = 10.45VIYY9 pKa = 9.16NTDD12 pKa = 3.37EE13 pKa = 4.19KK14 pKa = 11.26DD15 pKa = 3.71SNVFSARR22 pKa = 11.84STDD25 pKa = 3.37EE26 pKa = 4.14AFDD29 pKa = 3.66IAFQTIFDD37 pKa = 4.06TEE39 pKa = 4.12EE40 pKa = 3.6FDD42 pKa = 5.63EE43 pKa = 4.77FFNDD47 pKa = 3.19NGDD50 pKa = 3.66IVVTKK55 pKa = 10.61ANKK58 pKa = 10.2AYY60 pKa = 10.36FFEE63 pKa = 4.81CLKK66 pKa = 9.81GWKK69 pKa = 9.13MYY71 pKa = 10.73DD72 pKa = 3.22YY73 pKa = 11.45GKK75 pKa = 10.78YY76 pKa = 10.58DD77 pKa = 3.16SDD79 pKa = 3.68KK80 pKa = 11.21VLFSLTEE87 pKa = 3.85HH88 pKa = 7.16PNPIDD93 pKa = 3.38FNEE96 pKa = 4.03YY97 pKa = 10.86LKK99 pKa = 10.52ILQDD103 pKa = 3.58KK104 pKa = 10.66VVGLL108 pKa = 4.05
Molecular weight: 12.75 kDa
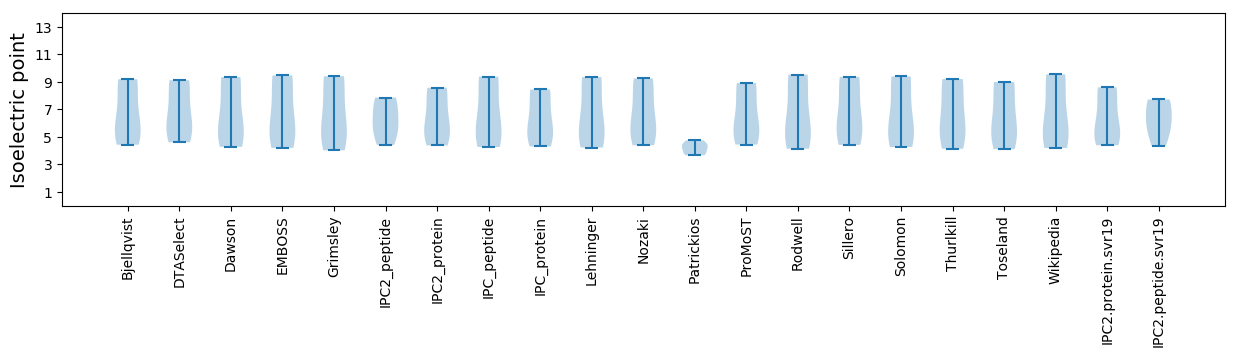
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W9I5|A0A4P8W9I5_9VIRU Replication initiation protein OS=Tortoise microvirus 26 OX=2583128 PE=4 SV=1
MM1 pKa = 7.19KK2 pKa = 10.04CSYY5 pKa = 10.5PFTVKK10 pKa = 10.6QNDD13 pKa = 2.84IHH15 pKa = 7.05GNNIYY20 pKa = 10.57QDD22 pKa = 3.95VACKK26 pKa = 10.16KK27 pKa = 10.37CYY29 pKa = 9.83SCRR32 pKa = 11.84KK33 pKa = 9.63SISQEE38 pKa = 3.09WFIRR42 pKa = 11.84LRR44 pKa = 11.84NEE46 pKa = 3.76LHH48 pKa = 5.25YY49 pKa = 10.72HH50 pKa = 4.88EE51 pKa = 5.99HH52 pKa = 6.33NSFVTLTYY60 pKa = 10.54SDD62 pKa = 3.76EE63 pKa = 4.33HH64 pKa = 7.76LPSDD68 pKa = 4.07FSISKK73 pKa = 10.4KK74 pKa = 10.4DD75 pKa = 3.26LQLFFKK81 pKa = 10.71RR82 pKa = 11.84LRR84 pKa = 11.84KK85 pKa = 9.32RR86 pKa = 11.84RR87 pKa = 11.84KK88 pKa = 9.39SKK90 pKa = 10.78LKK92 pKa = 10.05YY93 pKa = 9.12FACGEE98 pKa = 4.29YY99 pKa = 10.64GSQSYY104 pKa = 9.43RR105 pKa = 11.84PHH107 pKa = 5.41YY108 pKa = 10.2HH109 pKa = 7.08LILFGVGLYY118 pKa = 10.41SEE120 pKa = 5.35DD121 pKa = 3.47KK122 pKa = 11.1LDD124 pKa = 5.48IIEE127 pKa = 4.21SWPYY131 pKa = 11.19ADD133 pKa = 3.67WSVPSILKK141 pKa = 10.54NSFGLVEE148 pKa = 4.84DD149 pKa = 4.04KK150 pKa = 10.91SIKK153 pKa = 9.49YY154 pKa = 8.64VCNYY158 pKa = 9.12VNKK161 pKa = 10.33SLNDD165 pKa = 3.61LEE167 pKa = 4.44MKK169 pKa = 10.39IIYY172 pKa = 9.15QDD174 pKa = 3.6NNMTPPFRR182 pKa = 11.84IMSHH186 pKa = 5.8GLGLLYY192 pKa = 10.31MEE194 pKa = 5.41DD195 pKa = 3.95NKK197 pKa = 11.05TKK199 pKa = 10.85LIDD202 pKa = 3.58EE203 pKa = 4.15QVYY206 pKa = 8.46RR207 pKa = 11.84HH208 pKa = 5.8QGKK211 pKa = 9.77EE212 pKa = 3.82YY213 pKa = 10.21NLPYY217 pKa = 10.53YY218 pKa = 10.12YY219 pKa = 10.31RR220 pKa = 11.84NKK222 pKa = 10.59LNIGIKK228 pKa = 9.43TSPKK232 pKa = 10.01FQDD235 pKa = 2.96KK236 pKa = 11.03VIDD239 pKa = 3.79YY240 pKa = 8.66RR241 pKa = 11.84AKK243 pKa = 9.94QIKK246 pKa = 9.8EE247 pKa = 3.47ITGIDD252 pKa = 3.5AEE254 pKa = 4.77GEE256 pKa = 3.87ALEE259 pKa = 4.15YY260 pKa = 11.26LEE262 pKa = 5.58LDD264 pKa = 3.19SFVRR268 pKa = 11.84VRR270 pKa = 11.84KK271 pKa = 8.8HH272 pKa = 4.53QRR274 pKa = 11.84SDD276 pKa = 3.08QRR278 pKa = 11.84EE279 pKa = 3.78RR280 pKa = 11.84SLKK283 pKa = 10.51AKK285 pKa = 10.28QSLFNKK291 pKa = 10.42DD292 pKa = 2.88KK293 pKa = 10.97FF294 pKa = 3.83
MM1 pKa = 7.19KK2 pKa = 10.04CSYY5 pKa = 10.5PFTVKK10 pKa = 10.6QNDD13 pKa = 2.84IHH15 pKa = 7.05GNNIYY20 pKa = 10.57QDD22 pKa = 3.95VACKK26 pKa = 10.16KK27 pKa = 10.37CYY29 pKa = 9.83SCRR32 pKa = 11.84KK33 pKa = 9.63SISQEE38 pKa = 3.09WFIRR42 pKa = 11.84LRR44 pKa = 11.84NEE46 pKa = 3.76LHH48 pKa = 5.25YY49 pKa = 10.72HH50 pKa = 4.88EE51 pKa = 5.99HH52 pKa = 6.33NSFVTLTYY60 pKa = 10.54SDD62 pKa = 3.76EE63 pKa = 4.33HH64 pKa = 7.76LPSDD68 pKa = 4.07FSISKK73 pKa = 10.4KK74 pKa = 10.4DD75 pKa = 3.26LQLFFKK81 pKa = 10.71RR82 pKa = 11.84LRR84 pKa = 11.84KK85 pKa = 9.32RR86 pKa = 11.84RR87 pKa = 11.84KK88 pKa = 9.39SKK90 pKa = 10.78LKK92 pKa = 10.05YY93 pKa = 9.12FACGEE98 pKa = 4.29YY99 pKa = 10.64GSQSYY104 pKa = 9.43RR105 pKa = 11.84PHH107 pKa = 5.41YY108 pKa = 10.2HH109 pKa = 7.08LILFGVGLYY118 pKa = 10.41SEE120 pKa = 5.35DD121 pKa = 3.47KK122 pKa = 11.1LDD124 pKa = 5.48IIEE127 pKa = 4.21SWPYY131 pKa = 11.19ADD133 pKa = 3.67WSVPSILKK141 pKa = 10.54NSFGLVEE148 pKa = 4.84DD149 pKa = 4.04KK150 pKa = 10.91SIKK153 pKa = 9.49YY154 pKa = 8.64VCNYY158 pKa = 9.12VNKK161 pKa = 10.33SLNDD165 pKa = 3.61LEE167 pKa = 4.44MKK169 pKa = 10.39IIYY172 pKa = 9.15QDD174 pKa = 3.6NNMTPPFRR182 pKa = 11.84IMSHH186 pKa = 5.8GLGLLYY192 pKa = 10.31MEE194 pKa = 5.41DD195 pKa = 3.95NKK197 pKa = 11.05TKK199 pKa = 10.85LIDD202 pKa = 3.58EE203 pKa = 4.15QVYY206 pKa = 8.46RR207 pKa = 11.84HH208 pKa = 5.8QGKK211 pKa = 9.77EE212 pKa = 3.82YY213 pKa = 10.21NLPYY217 pKa = 10.53YY218 pKa = 10.12YY219 pKa = 10.31RR220 pKa = 11.84NKK222 pKa = 10.59LNIGIKK228 pKa = 9.43TSPKK232 pKa = 10.01FQDD235 pKa = 2.96KK236 pKa = 11.03VIDD239 pKa = 3.79YY240 pKa = 8.66RR241 pKa = 11.84AKK243 pKa = 9.94QIKK246 pKa = 9.8EE247 pKa = 3.47ITGIDD252 pKa = 3.5AEE254 pKa = 4.77GEE256 pKa = 3.87ALEE259 pKa = 4.15YY260 pKa = 11.26LEE262 pKa = 5.58LDD264 pKa = 3.19SFVRR268 pKa = 11.84VRR270 pKa = 11.84KK271 pKa = 8.8HH272 pKa = 4.53QRR274 pKa = 11.84SDD276 pKa = 3.08QRR278 pKa = 11.84EE279 pKa = 3.78RR280 pKa = 11.84SLKK283 pKa = 10.51AKK285 pKa = 10.28QSLFNKK291 pKa = 10.42DD292 pKa = 2.88KK293 pKa = 10.97FF294 pKa = 3.83
Molecular weight: 35.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1332 |
108 |
520 |
266.4 |
30.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.33 ± 1.224 | 0.976 ± 0.401 |
6.532 ± 0.641 | 5.405 ± 0.517 |
5.48 ± 0.712 | 6.006 ± 1.033 |
1.727 ± 0.608 | 5.931 ± 0.302 |
6.532 ± 1.418 | 8.108 ± 0.559 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.327 ± 0.275 | 6.006 ± 0.463 |
5.18 ± 0.905 | 5.856 ± 0.905 |
5.18 ± 0.428 | 7.808 ± 0.685 |
4.429 ± 1.071 | 4.429 ± 0.478 |
1.351 ± 0.446 | 5.405 ± 0.678 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
