
Tortoise microvirus 1
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.48
Get precalculated fractions of proteins
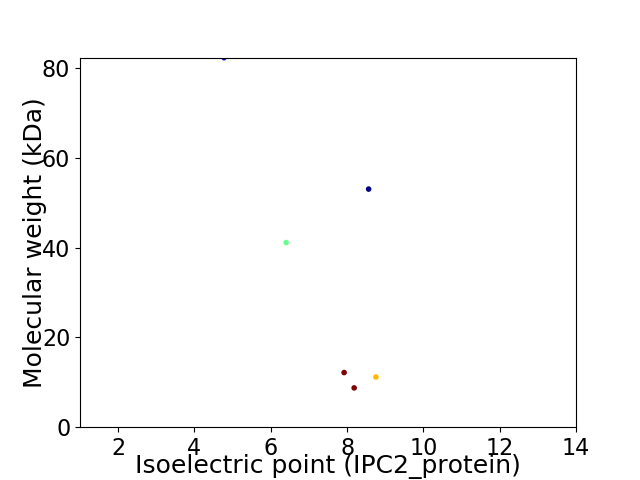
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W915|A0A4P8W915_9VIRU Replication initiation protein OS=Tortoise microvirus 1 OX=2583098 PE=4 SV=1
MM1 pKa = 7.0SQNIFDD7 pKa = 4.12ATFDD11 pKa = 3.85ANNQINVNSFDD22 pKa = 3.46WSHH25 pKa = 5.82VNNLTTSFGRR35 pKa = 11.84VTPVFCEE42 pKa = 3.92LVPAKK47 pKa = 10.42GSLRR51 pKa = 11.84INPEE55 pKa = 3.61FGLEE59 pKa = 3.97LMPMVFPVQTRR70 pKa = 11.84MYY72 pKa = 11.01ARR74 pKa = 11.84LNFFKK79 pKa = 10.48VTLRR83 pKa = 11.84SMWEE87 pKa = 4.14DD88 pKa = 3.36YY89 pKa = 11.61SDD91 pKa = 4.78FISNFRR97 pKa = 11.84DD98 pKa = 3.77DD99 pKa = 5.34LEE101 pKa = 4.37EE102 pKa = 4.61PYY104 pKa = 10.41ILPDD108 pKa = 3.71FVRR111 pKa = 11.84FAKK114 pKa = 9.32MCKK117 pKa = 9.11TGTLGDD123 pKa = 4.0YY124 pKa = 10.89LGLPTFGTNISQSVTIPDD142 pKa = 3.68SPCTVNSRR150 pKa = 11.84NFTTSVEE157 pKa = 4.22VNAAMASIRR166 pKa = 11.84QDD168 pKa = 3.09ATPAGLICANHH179 pKa = 6.37FNTDD183 pKa = 2.84GKK185 pKa = 10.11GTVIFKK191 pKa = 6.96TQRR194 pKa = 11.84SATQIPQSVSSVGINIGIVGSSADD218 pKa = 3.44AFVGCRR224 pKa = 11.84GYY226 pKa = 10.76LVHH229 pKa = 7.22YY230 pKa = 9.92GDD232 pKa = 5.52DD233 pKa = 3.35GTYY236 pKa = 9.62YY237 pKa = 10.5SDD239 pKa = 4.14YY240 pKa = 9.15PVEE243 pKa = 4.39VIKK246 pKa = 11.13SDD248 pKa = 3.73DD249 pKa = 3.66GVVGVVISSIDD260 pKa = 3.05IGSFSNALGGLDD272 pKa = 4.27LVLNAPNSANPIEE285 pKa = 4.37FTGMQYY291 pKa = 11.14NVVVSYY297 pKa = 8.34YY298 pKa = 9.66TLQSDD303 pKa = 3.99VSYY306 pKa = 10.43TKK308 pKa = 11.02YY309 pKa = 10.34PFATEE314 pKa = 3.53EE315 pKa = 4.06HH316 pKa = 6.82LNFPRR321 pKa = 11.84LLAYY325 pKa = 10.09RR326 pKa = 11.84FRR328 pKa = 11.84AYY330 pKa = 10.2EE331 pKa = 4.04SVYY334 pKa = 9.54NAYY337 pKa = 10.51YY338 pKa = 10.44RR339 pKa = 11.84DD340 pKa = 3.2IRR342 pKa = 11.84NNPFVINGRR351 pKa = 11.84PVYY354 pKa = 10.54NKK356 pKa = 9.03WLPNMKK362 pKa = 10.16GGADD366 pKa = 3.25TTLYY370 pKa = 10.46EE371 pKa = 3.95LHH373 pKa = 5.6QCNWEE378 pKa = 3.97KK379 pKa = 11.33DD380 pKa = 3.84FLTTAVPNPQQGTNPPLVGLTVGDD404 pKa = 3.96VVTRR408 pKa = 11.84TSDD411 pKa = 2.99GTLQVQKK418 pKa = 9.47QTVLVDD424 pKa = 3.41EE425 pKa = 5.81DD426 pKa = 3.95GAKK429 pKa = 10.61YY430 pKa = 10.13GISYY434 pKa = 10.01KK435 pKa = 10.42VSEE438 pKa = 5.18DD439 pKa = 3.46GQSLVGVDD447 pKa = 3.92YY448 pKa = 11.39DD449 pKa = 3.9PVSDD453 pKa = 4.28KK454 pKa = 11.43TPITQINSYY463 pKa = 11.33AEE465 pKa = 4.17LASLATEE472 pKa = 3.97QGSGFTIEE480 pKa = 3.8TLRR483 pKa = 11.84YY484 pKa = 8.15VNAYY488 pKa = 10.02QKK490 pKa = 10.64FLEE493 pKa = 4.19LNMRR497 pKa = 11.84KK498 pKa = 9.68GFSYY502 pKa = 9.93KK503 pKa = 10.37QIMQGRR509 pKa = 11.84WDD511 pKa = 3.19INIRR515 pKa = 11.84FDD517 pKa = 3.83EE518 pKa = 4.39LLMPEE523 pKa = 4.69FIGGVSRR530 pKa = 11.84EE531 pKa = 3.73LSMRR535 pKa = 11.84TVEE538 pKa = 4.11QTVDD542 pKa = 3.26QQTEE546 pKa = 3.88ISQGQYY552 pKa = 11.21AEE554 pKa = 4.34ALGSKK559 pKa = 8.88TGIAGVYY566 pKa = 10.3GYY568 pKa = 11.41GNGNIEE574 pKa = 4.2VFCDD578 pKa = 3.47EE579 pKa = 4.08EE580 pKa = 5.21SIIIGLLTVTPVPVYY595 pKa = 9.03TQLMPKK601 pKa = 10.27DD602 pKa = 3.52FLYY605 pKa = 11.18NGLLDD610 pKa = 4.32HH611 pKa = 6.36YY612 pKa = 10.57QPEE615 pKa = 4.21FDD617 pKa = 5.15RR618 pKa = 11.84IGFQPITYY626 pKa = 10.11KK627 pKa = 10.3EE628 pKa = 4.25VCPMNIPAYY637 pKa = 10.41DD638 pKa = 3.61DD639 pKa = 3.78TNQLEE644 pKa = 4.38QTFGYY649 pKa = 8.8QRR651 pKa = 11.84PWYY654 pKa = 9.05EE655 pKa = 3.53YY656 pKa = 8.47VAKK659 pKa = 10.41YY660 pKa = 10.67DD661 pKa = 3.64SAHH664 pKa = 5.62GLFRR668 pKa = 11.84TSMKK672 pKa = 10.31NFIMSRR678 pKa = 11.84VFRR681 pKa = 11.84GLPQLGQEE689 pKa = 4.52FLLVDD694 pKa = 4.51PNTTNQVFAVTEE706 pKa = 4.01YY707 pKa = 10.13SDD709 pKa = 4.98KK710 pKa = 10.98IFGYY714 pKa = 10.82VKK716 pKa = 10.25FNATARR722 pKa = 11.84LPISRR727 pKa = 11.84VAIPRR732 pKa = 11.84LDD734 pKa = 3.18
MM1 pKa = 7.0SQNIFDD7 pKa = 4.12ATFDD11 pKa = 3.85ANNQINVNSFDD22 pKa = 3.46WSHH25 pKa = 5.82VNNLTTSFGRR35 pKa = 11.84VTPVFCEE42 pKa = 3.92LVPAKK47 pKa = 10.42GSLRR51 pKa = 11.84INPEE55 pKa = 3.61FGLEE59 pKa = 3.97LMPMVFPVQTRR70 pKa = 11.84MYY72 pKa = 11.01ARR74 pKa = 11.84LNFFKK79 pKa = 10.48VTLRR83 pKa = 11.84SMWEE87 pKa = 4.14DD88 pKa = 3.36YY89 pKa = 11.61SDD91 pKa = 4.78FISNFRR97 pKa = 11.84DD98 pKa = 3.77DD99 pKa = 5.34LEE101 pKa = 4.37EE102 pKa = 4.61PYY104 pKa = 10.41ILPDD108 pKa = 3.71FVRR111 pKa = 11.84FAKK114 pKa = 9.32MCKK117 pKa = 9.11TGTLGDD123 pKa = 4.0YY124 pKa = 10.89LGLPTFGTNISQSVTIPDD142 pKa = 3.68SPCTVNSRR150 pKa = 11.84NFTTSVEE157 pKa = 4.22VNAAMASIRR166 pKa = 11.84QDD168 pKa = 3.09ATPAGLICANHH179 pKa = 6.37FNTDD183 pKa = 2.84GKK185 pKa = 10.11GTVIFKK191 pKa = 6.96TQRR194 pKa = 11.84SATQIPQSVSSVGINIGIVGSSADD218 pKa = 3.44AFVGCRR224 pKa = 11.84GYY226 pKa = 10.76LVHH229 pKa = 7.22YY230 pKa = 9.92GDD232 pKa = 5.52DD233 pKa = 3.35GTYY236 pKa = 9.62YY237 pKa = 10.5SDD239 pKa = 4.14YY240 pKa = 9.15PVEE243 pKa = 4.39VIKK246 pKa = 11.13SDD248 pKa = 3.73DD249 pKa = 3.66GVVGVVISSIDD260 pKa = 3.05IGSFSNALGGLDD272 pKa = 4.27LVLNAPNSANPIEE285 pKa = 4.37FTGMQYY291 pKa = 11.14NVVVSYY297 pKa = 8.34YY298 pKa = 9.66TLQSDD303 pKa = 3.99VSYY306 pKa = 10.43TKK308 pKa = 11.02YY309 pKa = 10.34PFATEE314 pKa = 3.53EE315 pKa = 4.06HH316 pKa = 6.82LNFPRR321 pKa = 11.84LLAYY325 pKa = 10.09RR326 pKa = 11.84FRR328 pKa = 11.84AYY330 pKa = 10.2EE331 pKa = 4.04SVYY334 pKa = 9.54NAYY337 pKa = 10.51YY338 pKa = 10.44RR339 pKa = 11.84DD340 pKa = 3.2IRR342 pKa = 11.84NNPFVINGRR351 pKa = 11.84PVYY354 pKa = 10.54NKK356 pKa = 9.03WLPNMKK362 pKa = 10.16GGADD366 pKa = 3.25TTLYY370 pKa = 10.46EE371 pKa = 3.95LHH373 pKa = 5.6QCNWEE378 pKa = 3.97KK379 pKa = 11.33DD380 pKa = 3.84FLTTAVPNPQQGTNPPLVGLTVGDD404 pKa = 3.96VVTRR408 pKa = 11.84TSDD411 pKa = 2.99GTLQVQKK418 pKa = 9.47QTVLVDD424 pKa = 3.41EE425 pKa = 5.81DD426 pKa = 3.95GAKK429 pKa = 10.61YY430 pKa = 10.13GISYY434 pKa = 10.01KK435 pKa = 10.42VSEE438 pKa = 5.18DD439 pKa = 3.46GQSLVGVDD447 pKa = 3.92YY448 pKa = 11.39DD449 pKa = 3.9PVSDD453 pKa = 4.28KK454 pKa = 11.43TPITQINSYY463 pKa = 11.33AEE465 pKa = 4.17LASLATEE472 pKa = 3.97QGSGFTIEE480 pKa = 3.8TLRR483 pKa = 11.84YY484 pKa = 8.15VNAYY488 pKa = 10.02QKK490 pKa = 10.64FLEE493 pKa = 4.19LNMRR497 pKa = 11.84KK498 pKa = 9.68GFSYY502 pKa = 9.93KK503 pKa = 10.37QIMQGRR509 pKa = 11.84WDD511 pKa = 3.19INIRR515 pKa = 11.84FDD517 pKa = 3.83EE518 pKa = 4.39LLMPEE523 pKa = 4.69FIGGVSRR530 pKa = 11.84EE531 pKa = 3.73LSMRR535 pKa = 11.84TVEE538 pKa = 4.11QTVDD542 pKa = 3.26QQTEE546 pKa = 3.88ISQGQYY552 pKa = 11.21AEE554 pKa = 4.34ALGSKK559 pKa = 8.88TGIAGVYY566 pKa = 10.3GYY568 pKa = 11.41GNGNIEE574 pKa = 4.2VFCDD578 pKa = 3.47EE579 pKa = 4.08EE580 pKa = 5.21SIIIGLLTVTPVPVYY595 pKa = 9.03TQLMPKK601 pKa = 10.27DD602 pKa = 3.52FLYY605 pKa = 11.18NGLLDD610 pKa = 4.32HH611 pKa = 6.36YY612 pKa = 10.57QPEE615 pKa = 4.21FDD617 pKa = 5.15RR618 pKa = 11.84IGFQPITYY626 pKa = 10.11KK627 pKa = 10.3EE628 pKa = 4.25VCPMNIPAYY637 pKa = 10.41DD638 pKa = 3.61DD639 pKa = 3.78TNQLEE644 pKa = 4.38QTFGYY649 pKa = 8.8QRR651 pKa = 11.84PWYY654 pKa = 9.05EE655 pKa = 3.53YY656 pKa = 8.47VAKK659 pKa = 10.41YY660 pKa = 10.67DD661 pKa = 3.64SAHH664 pKa = 5.62GLFRR668 pKa = 11.84TSMKK672 pKa = 10.31NFIMSRR678 pKa = 11.84VFRR681 pKa = 11.84GLPQLGQEE689 pKa = 4.52FLLVDD694 pKa = 4.51PNTTNQVFAVTEE706 pKa = 4.01YY707 pKa = 10.13SDD709 pKa = 4.98KK710 pKa = 10.98IFGYY714 pKa = 10.82VKK716 pKa = 10.25FNATARR722 pKa = 11.84LPISRR727 pKa = 11.84VAIPRR732 pKa = 11.84LDD734 pKa = 3.18
Molecular weight: 82.28 kDa
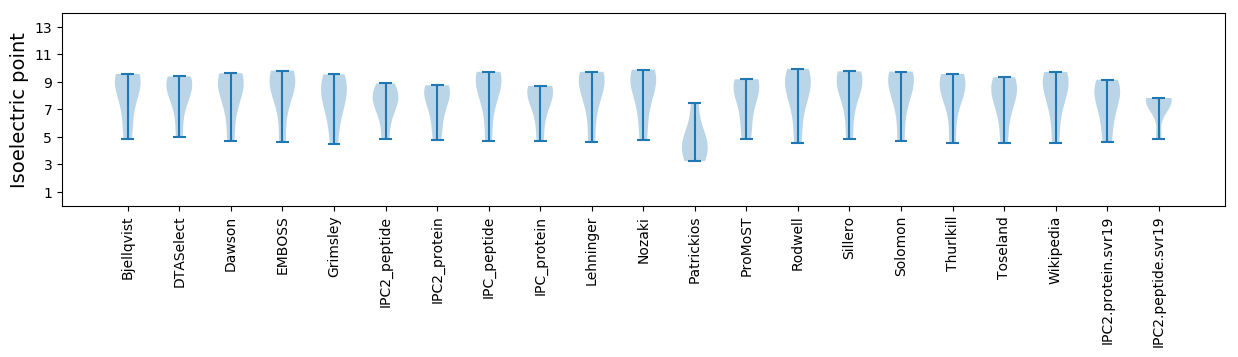
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVV8|A0A4V1FVV8_9VIRU Uncharacterized protein OS=Tortoise microvirus 1 OX=2583098 PE=4 SV=1
MM1 pKa = 7.73LLQLWCEE8 pKa = 3.85NNFFFFLPTLHH19 pKa = 6.98SLAFCVSCSKK29 pKa = 10.78GYY31 pKa = 9.12SVKK34 pKa = 10.16FCYY37 pKa = 10.4NFGACGKK44 pKa = 9.98ILIKK48 pKa = 10.54FNGMFIGLLTQNILSLQFRR67 pKa = 11.84KK68 pKa = 10.04IKK70 pKa = 10.29GKK72 pKa = 10.62SIFPLRR78 pKa = 11.84EE79 pKa = 3.53TQINSNFVACVRR91 pKa = 11.84DD92 pKa = 3.82CRR94 pKa = 11.84RR95 pKa = 11.84VSS97 pKa = 3.02
MM1 pKa = 7.73LLQLWCEE8 pKa = 3.85NNFFFFLPTLHH19 pKa = 6.98SLAFCVSCSKK29 pKa = 10.78GYY31 pKa = 9.12SVKK34 pKa = 10.16FCYY37 pKa = 10.4NFGACGKK44 pKa = 9.98ILIKK48 pKa = 10.54FNGMFIGLLTQNILSLQFRR67 pKa = 11.84KK68 pKa = 10.04IKK70 pKa = 10.29GKK72 pKa = 10.62SIFPLRR78 pKa = 11.84EE79 pKa = 3.53TQINSNFVACVRR91 pKa = 11.84DD92 pKa = 3.82CRR94 pKa = 11.84RR95 pKa = 11.84VSS97 pKa = 3.02
Molecular weight: 11.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1842 |
75 |
734 |
307.0 |
34.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.003 ± 1.528 | 1.9 ± 0.749 |
5.049 ± 0.587 | 4.832 ± 0.401 |
5.646 ± 0.917 | 6.026 ± 0.861 |
1.466 ± 0.678 | 5.809 ± 0.369 |
5.809 ± 1.096 | 7.329 ± 0.428 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.769 ± 0.312 | 6.515 ± 0.347 |
4.669 ± 0.478 | 4.94 ± 0.952 |
4.669 ± 0.21 | 7.275 ± 0.328 |
5.537 ± 0.858 | 6.406 ± 0.95 |
0.869 ± 0.127 | 5.483 ± 1.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
