
Lichenicoccus roseus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Lichenicoccus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
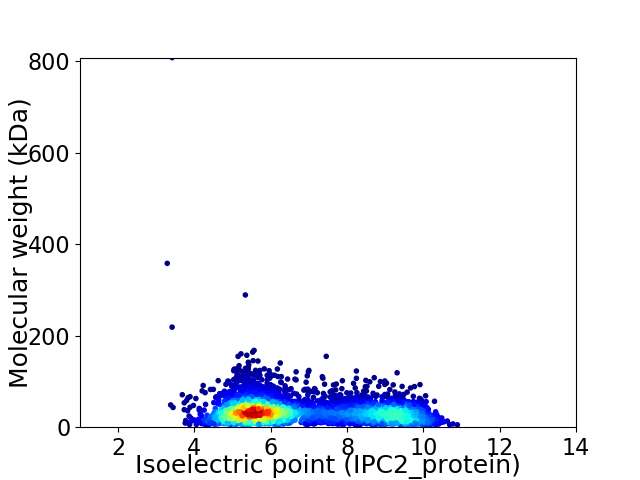
Virtual 2D-PAGE plot for 4254 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5R9J600|A0A5R9J600_9PROT Uncharacterized protein OS=Lichenicoccus roseus OX=2683649 GN=FE263_09950 PE=4 SV=1
MM1 pKa = 7.17TAFSALSAAAISTQTIATIAALTTADD27 pKa = 4.51FAPLTAQQMDD37 pKa = 4.33ALTSAQWAAIVPAALSALTTTQVYY61 pKa = 8.67TISSTALAGLTSAQLVAMSSTSVEE85 pKa = 4.08ALEE88 pKa = 5.21GPQIPGLTIAALAGLSDD105 pKa = 3.89AEE107 pKa = 3.85IAGFSEE113 pKa = 3.84QFMYY117 pKa = 8.99ITPAQLASLSIDD129 pKa = 3.11QFQAISSNSLLLLTASQFGAFTTTQIQALTNAQLIGLSGKK169 pKa = 9.31QCAYY173 pKa = 11.0LSGPQLAAVSTANLAALTAYY193 pKa = 9.36QAQSISVAALASLTSTQVAALTPAILASLAGKK225 pKa = 9.85SLAAISNAALAGATTDD241 pKa = 4.34EE242 pKa = 4.36IAGLAGMGQFATLTVAQIAALSPVQLGAITTFEE275 pKa = 4.01MTGITTAQLQALTAAQIAYY294 pKa = 7.54LTPAAFATLGSAQLHH309 pKa = 5.96ALSGAQLNAVSSTDD323 pKa = 3.07IASISSATIAGLSTAALAGLGSTQIDD349 pKa = 3.96GLDD352 pKa = 3.01IAFAGVTTAQLASLTSTTLASLDD375 pKa = 3.41GRR377 pKa = 11.84EE378 pKa = 4.15WASLTTSQIASFTTAQIASLAPAALAAIPTANYY411 pKa = 10.57AGLTYY416 pKa = 10.95ADD418 pKa = 3.43IAALTTAQVAALVPSEE434 pKa = 3.94ILSFSEE440 pKa = 3.98LQIASLTKK448 pKa = 9.64PQVASISITDD458 pKa = 3.8IAALGTDD465 pKa = 4.17PARR468 pKa = 11.84MLSTNAIAGLTATQIVAMASTSVEE492 pKa = 3.98ALTEE496 pKa = 4.09NQVPALSLATFDD508 pKa = 4.39ALTPAEE514 pKa = 3.97LAGFVNNQIAFMTVAEE530 pKa = 4.37VASMSVLQIQALGSNLLTQLTAPQLAALTTAQIAALTTTQVAGLGSTQFAAFSAPQAAALNLADD594 pKa = 5.44LAGLTTNQATSLSLAALAALTSTQVAAISSSALEE628 pKa = 4.02ALSARR633 pKa = 11.84GIAAITTAALAGVSTDD649 pKa = 3.87EE650 pKa = 4.23IAALTAYY657 pKa = 10.52ASGQFQALTAAQIAALSTAQVAAITSGEE685 pKa = 4.09VLGITSAQLQALTPRR700 pKa = 11.84EE701 pKa = 3.91IAALTSVQAGTMTSTQLL718 pKa = 3.3
MM1 pKa = 7.17TAFSALSAAAISTQTIATIAALTTADD27 pKa = 4.51FAPLTAQQMDD37 pKa = 4.33ALTSAQWAAIVPAALSALTTTQVYY61 pKa = 8.67TISSTALAGLTSAQLVAMSSTSVEE85 pKa = 4.08ALEE88 pKa = 5.21GPQIPGLTIAALAGLSDD105 pKa = 3.89AEE107 pKa = 3.85IAGFSEE113 pKa = 3.84QFMYY117 pKa = 8.99ITPAQLASLSIDD129 pKa = 3.11QFQAISSNSLLLLTASQFGAFTTTQIQALTNAQLIGLSGKK169 pKa = 9.31QCAYY173 pKa = 11.0LSGPQLAAVSTANLAALTAYY193 pKa = 9.36QAQSISVAALASLTSTQVAALTPAILASLAGKK225 pKa = 9.85SLAAISNAALAGATTDD241 pKa = 4.34EE242 pKa = 4.36IAGLAGMGQFATLTVAQIAALSPVQLGAITTFEE275 pKa = 4.01MTGITTAQLQALTAAQIAYY294 pKa = 7.54LTPAAFATLGSAQLHH309 pKa = 5.96ALSGAQLNAVSSTDD323 pKa = 3.07IASISSATIAGLSTAALAGLGSTQIDD349 pKa = 3.96GLDD352 pKa = 3.01IAFAGVTTAQLASLTSTTLASLDD375 pKa = 3.41GRR377 pKa = 11.84EE378 pKa = 4.15WASLTTSQIASFTTAQIASLAPAALAAIPTANYY411 pKa = 10.57AGLTYY416 pKa = 10.95ADD418 pKa = 3.43IAALTTAQVAALVPSEE434 pKa = 3.94ILSFSEE440 pKa = 3.98LQIASLTKK448 pKa = 9.64PQVASISITDD458 pKa = 3.8IAALGTDD465 pKa = 4.17PARR468 pKa = 11.84MLSTNAIAGLTATQIVAMASTSVEE492 pKa = 3.98ALTEE496 pKa = 4.09NQVPALSLATFDD508 pKa = 4.39ALTPAEE514 pKa = 3.97LAGFVNNQIAFMTVAEE530 pKa = 4.37VASMSVLQIQALGSNLLTQLTAPQLAALTTAQIAALTTTQVAGLGSTQFAAFSAPQAAALNLADD594 pKa = 5.44LAGLTTNQATSLSLAALAALTSTQVAAISSSALEE628 pKa = 4.02ALSARR633 pKa = 11.84GIAAITTAALAGVSTDD649 pKa = 3.87EE650 pKa = 4.23IAALTAYY657 pKa = 10.52ASGQFQALTAAQIAALSTAQVAAITSGEE685 pKa = 4.09VLGITSAQLQALTPRR700 pKa = 11.84EE701 pKa = 3.91IAALTSVQAGTMTSTQLL718 pKa = 3.3
Molecular weight: 70.95 kDa
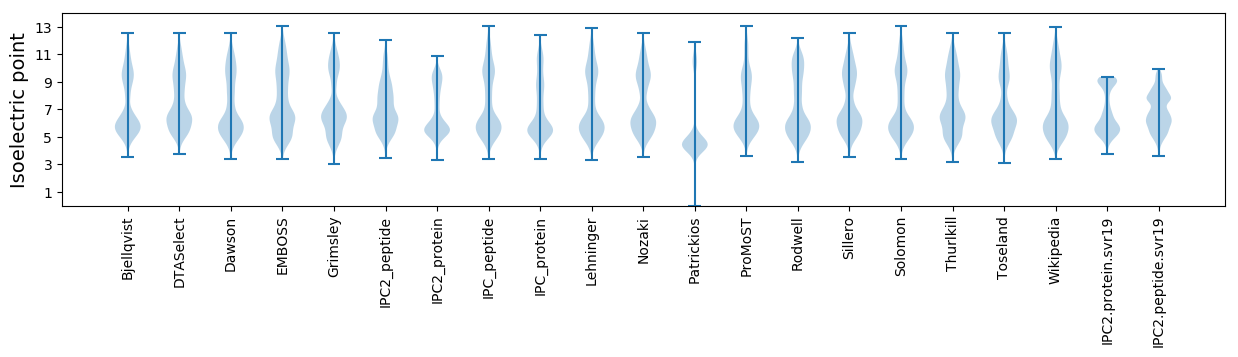
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5R9J9H8|A0A5R9J9H8_9PROT SDR family oxidoreductase OS=Lichenicoccus roseus OX=2683649 GN=FE263_03185 PE=4 SV=1
MM1 pKa = 7.39RR2 pKa = 11.84TLLTLLMVFAMIGTVGVLAAGVVGFVRR29 pKa = 11.84GGGDD33 pKa = 3.16PRR35 pKa = 11.84RR36 pKa = 11.84SNRR39 pKa = 11.84LMRR42 pKa = 11.84WRR44 pKa = 11.84VILQASAILLFVLLLTIFRR63 pKa = 11.84RR64 pKa = 3.89
MM1 pKa = 7.39RR2 pKa = 11.84TLLTLLMVFAMIGTVGVLAAGVVGFVRR29 pKa = 11.84GGGDD33 pKa = 3.16PRR35 pKa = 11.84RR36 pKa = 11.84SNRR39 pKa = 11.84LMRR42 pKa = 11.84WRR44 pKa = 11.84VILQASAILLFVLLLTIFRR63 pKa = 11.84RR64 pKa = 3.89
Molecular weight: 7.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1424553 |
26 |
8351 |
334.9 |
35.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.381 ± 0.051 | 0.925 ± 0.012 |
5.543 ± 0.029 | 4.793 ± 0.044 |
3.214 ± 0.025 | 9.18 ± 0.042 |
2.315 ± 0.019 | 4.606 ± 0.029 |
1.877 ± 0.028 | 11.037 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.31 ± 0.02 | 2.186 ± 0.023 |
5.707 ± 0.032 | 3.292 ± 0.028 |
7.898 ± 0.063 | 5.528 ± 0.048 |
5.373 ± 0.083 | 7.478 ± 0.03 |
1.378 ± 0.015 | 1.978 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
