
Inquilinus limosus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Inquilinus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
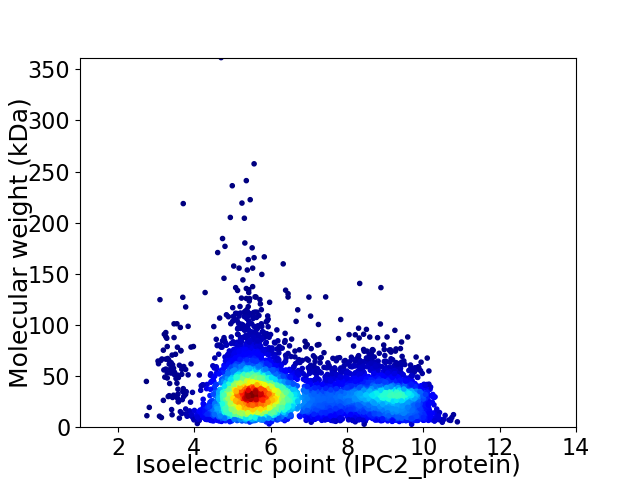
Virtual 2D-PAGE plot for 7046 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A211Z046|A0A211Z046_9PROT Aspartate-semialdehyde dehydrogenase OS=Inquilinus limosus OX=171674 GN=asd PE=3 SV=1
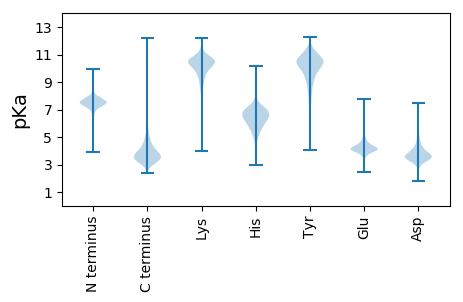
MM1 pKa = 7.66PLISNIAYY9 pKa = 10.47SPDD12 pKa = 3.61PTSGQALGLDD22 pKa = 3.73VALNGATRR30 pKa = 11.84LVVNWGDD37 pKa = 3.66GSPLQLYY44 pKa = 8.18QVEE47 pKa = 4.63PGSSSIHH54 pKa = 6.51LSHH57 pKa = 7.31AYY59 pKa = 10.17ADD61 pKa = 3.9PTVGTVVLRR70 pKa = 11.84AGGPGVPDD78 pKa = 4.65DD79 pKa = 4.1RR80 pKa = 11.84ATLNLVVGADD90 pKa = 3.55EE91 pKa = 4.56SYY93 pKa = 11.39SRR95 pKa = 11.84VSGDD99 pKa = 3.58PEE101 pKa = 4.18FDD103 pKa = 3.48TILRR107 pKa = 11.84EE108 pKa = 3.99NPLISGDD115 pKa = 3.72GNLVAWADD123 pKa = 4.16TINGHH128 pKa = 5.89SVFAMVKK135 pKa = 9.68NLATGTTDD143 pKa = 3.23YY144 pKa = 11.35LGVGRR149 pKa = 11.84DD150 pKa = 3.5AYY152 pKa = 11.05QFDD155 pKa = 4.69GPPVMLANGAAVWVYY170 pKa = 10.77EE171 pKa = 4.64GYY173 pKa = 10.8LGIAGYY179 pKa = 8.58LHH181 pKa = 7.02PASLYY186 pKa = 10.55LAATGQEE193 pKa = 3.78ITLEE197 pKa = 4.16DD198 pKa = 4.28AYY200 pKa = 10.74HH201 pKa = 5.8ITVTDD206 pKa = 3.76DD207 pKa = 2.9AKK209 pKa = 10.92YY210 pKa = 10.65AVYY213 pKa = 10.19QYY215 pKa = 10.01DD216 pKa = 3.79TEE218 pKa = 4.78LGSGQNGWYY227 pKa = 10.49LRR229 pKa = 11.84DD230 pKa = 3.54LDD232 pKa = 4.97AGTSVYY238 pKa = 10.69LGADD242 pKa = 3.51SEE244 pKa = 4.82YY245 pKa = 10.9DD246 pKa = 3.21GPRR249 pKa = 11.84YY250 pKa = 9.21HH251 pKa = 6.64VHH253 pKa = 6.61FVDD256 pKa = 4.12GQLVVDD262 pKa = 5.2DD263 pKa = 5.76DD264 pKa = 4.34STGQQSVSGALATDD278 pKa = 4.12TTADD282 pKa = 3.31GSVTVIATDD291 pKa = 3.65DD292 pKa = 3.61ALAPGDD298 pKa = 4.11TNNGSDD304 pKa = 4.62VYY306 pKa = 10.6LQEE309 pKa = 4.63NGSPTHH315 pKa = 7.29DD316 pKa = 3.31GTAGPDD322 pKa = 3.54VLLLGAANDD331 pKa = 3.87IADD334 pKa = 3.96GGAGKK339 pKa = 10.35DD340 pKa = 3.82YY341 pKa = 10.97LAGGAGDD348 pKa = 4.58DD349 pKa = 4.32SLSGGAGDD357 pKa = 5.54DD358 pKa = 3.99LLEE361 pKa = 4.9GGAGADD367 pKa = 3.9SLNGGDD373 pKa = 4.88GIDD376 pKa = 3.41TAGYY380 pKa = 7.28ATSLAGVQASLDD392 pKa = 3.66RR393 pKa = 11.84PAGNTGAAAGDD404 pKa = 3.95SYY406 pKa = 12.06AGIEE410 pKa = 4.15NLLGSAFADD419 pKa = 3.64RR420 pKa = 11.84LEE422 pKa = 5.19GDD424 pKa = 3.34AGANALRR431 pKa = 11.84GGAGDD436 pKa = 4.41DD437 pKa = 3.42RR438 pKa = 11.84LGGLGGDD445 pKa = 3.98DD446 pKa = 3.64RR447 pKa = 11.84LVGGDD452 pKa = 3.61GGDD455 pKa = 3.65IISGGSGNDD464 pKa = 3.33RR465 pKa = 11.84IYY467 pKa = 11.29GGAGDD472 pKa = 5.03DD473 pKa = 4.25RR474 pKa = 11.84MTGGAGADD482 pKa = 3.27VFVIQPGFGRR492 pKa = 11.84DD493 pKa = 3.24VVTDD497 pKa = 4.53FSHH500 pKa = 7.39AAGDD504 pKa = 3.85VILFDD509 pKa = 4.21RR510 pKa = 11.84DD511 pKa = 3.35VFASFAEE518 pKa = 4.67VMAHH522 pKa = 6.13AADD525 pKa = 3.92YY526 pKa = 10.57GADD529 pKa = 3.61GTVIALDD536 pKa = 4.3ADD538 pKa = 4.07DD539 pKa = 5.69KK540 pKa = 11.67LILLGVDD547 pKa = 3.15RR548 pKa = 11.84ASLVAGDD555 pKa = 3.78FAFVV559 pKa = 3.08
MM1 pKa = 7.66PLISNIAYY9 pKa = 10.47SPDD12 pKa = 3.61PTSGQALGLDD22 pKa = 3.73VALNGATRR30 pKa = 11.84LVVNWGDD37 pKa = 3.66GSPLQLYY44 pKa = 8.18QVEE47 pKa = 4.63PGSSSIHH54 pKa = 6.51LSHH57 pKa = 7.31AYY59 pKa = 10.17ADD61 pKa = 3.9PTVGTVVLRR70 pKa = 11.84AGGPGVPDD78 pKa = 4.65DD79 pKa = 4.1RR80 pKa = 11.84ATLNLVVGADD90 pKa = 3.55EE91 pKa = 4.56SYY93 pKa = 11.39SRR95 pKa = 11.84VSGDD99 pKa = 3.58PEE101 pKa = 4.18FDD103 pKa = 3.48TILRR107 pKa = 11.84EE108 pKa = 3.99NPLISGDD115 pKa = 3.72GNLVAWADD123 pKa = 4.16TINGHH128 pKa = 5.89SVFAMVKK135 pKa = 9.68NLATGTTDD143 pKa = 3.23YY144 pKa = 11.35LGVGRR149 pKa = 11.84DD150 pKa = 3.5AYY152 pKa = 11.05QFDD155 pKa = 4.69GPPVMLANGAAVWVYY170 pKa = 10.77EE171 pKa = 4.64GYY173 pKa = 10.8LGIAGYY179 pKa = 8.58LHH181 pKa = 7.02PASLYY186 pKa = 10.55LAATGQEE193 pKa = 3.78ITLEE197 pKa = 4.16DD198 pKa = 4.28AYY200 pKa = 10.74HH201 pKa = 5.8ITVTDD206 pKa = 3.76DD207 pKa = 2.9AKK209 pKa = 10.92YY210 pKa = 10.65AVYY213 pKa = 10.19QYY215 pKa = 10.01DD216 pKa = 3.79TEE218 pKa = 4.78LGSGQNGWYY227 pKa = 10.49LRR229 pKa = 11.84DD230 pKa = 3.54LDD232 pKa = 4.97AGTSVYY238 pKa = 10.69LGADD242 pKa = 3.51SEE244 pKa = 4.82YY245 pKa = 10.9DD246 pKa = 3.21GPRR249 pKa = 11.84YY250 pKa = 9.21HH251 pKa = 6.64VHH253 pKa = 6.61FVDD256 pKa = 4.12GQLVVDD262 pKa = 5.2DD263 pKa = 5.76DD264 pKa = 4.34STGQQSVSGALATDD278 pKa = 4.12TTADD282 pKa = 3.31GSVTVIATDD291 pKa = 3.65DD292 pKa = 3.61ALAPGDD298 pKa = 4.11TNNGSDD304 pKa = 4.62VYY306 pKa = 10.6LQEE309 pKa = 4.63NGSPTHH315 pKa = 7.29DD316 pKa = 3.31GTAGPDD322 pKa = 3.54VLLLGAANDD331 pKa = 3.87IADD334 pKa = 3.96GGAGKK339 pKa = 10.35DD340 pKa = 3.82YY341 pKa = 10.97LAGGAGDD348 pKa = 4.58DD349 pKa = 4.32SLSGGAGDD357 pKa = 5.54DD358 pKa = 3.99LLEE361 pKa = 4.9GGAGADD367 pKa = 3.9SLNGGDD373 pKa = 4.88GIDD376 pKa = 3.41TAGYY380 pKa = 7.28ATSLAGVQASLDD392 pKa = 3.66RR393 pKa = 11.84PAGNTGAAAGDD404 pKa = 3.95SYY406 pKa = 12.06AGIEE410 pKa = 4.15NLLGSAFADD419 pKa = 3.64RR420 pKa = 11.84LEE422 pKa = 5.19GDD424 pKa = 3.34AGANALRR431 pKa = 11.84GGAGDD436 pKa = 4.41DD437 pKa = 3.42RR438 pKa = 11.84LGGLGGDD445 pKa = 3.98DD446 pKa = 3.64RR447 pKa = 11.84LVGGDD452 pKa = 3.61GGDD455 pKa = 3.65IISGGSGNDD464 pKa = 3.33RR465 pKa = 11.84IYY467 pKa = 11.29GGAGDD472 pKa = 5.03DD473 pKa = 4.25RR474 pKa = 11.84MTGGAGADD482 pKa = 3.27VFVIQPGFGRR492 pKa = 11.84DD493 pKa = 3.24VVTDD497 pKa = 4.53FSHH500 pKa = 7.39AAGDD504 pKa = 3.85VILFDD509 pKa = 4.21RR510 pKa = 11.84DD511 pKa = 3.35VFASFAEE518 pKa = 4.67VMAHH522 pKa = 6.13AADD525 pKa = 3.92YY526 pKa = 10.57GADD529 pKa = 3.61GTVIALDD536 pKa = 4.3ADD538 pKa = 4.07DD539 pKa = 5.69KK540 pKa = 11.67LILLGVDD547 pKa = 3.15RR548 pKa = 11.84ASLVAGDD555 pKa = 3.78FAFVV559 pKa = 3.08
Molecular weight: 56.57 kDa
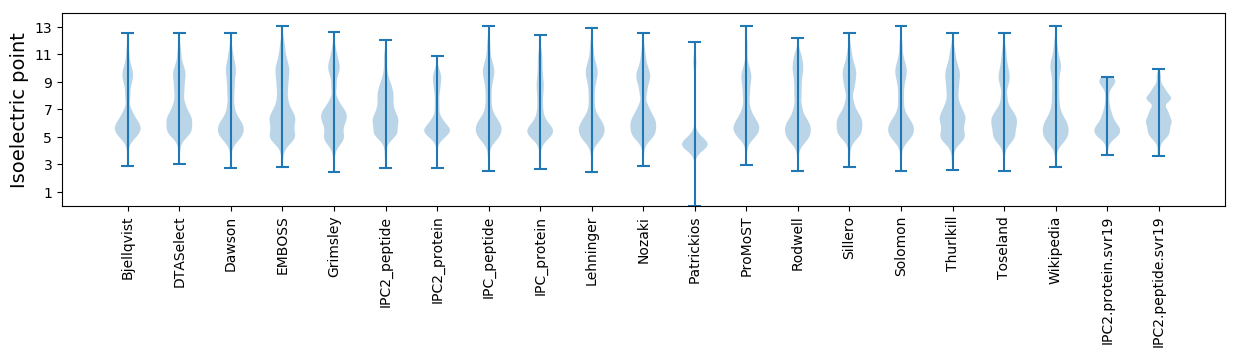
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A211ZIN8|A0A211ZIN8_9PROT MFS transporter OS=Inquilinus limosus OX=171674 GN=BWR60_20960 PE=4 SV=1
MM1 pKa = 7.2PRR3 pKa = 11.84EE4 pKa = 3.92RR5 pKa = 11.84QRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84ATVCVVSVSSQAMTSASRR27 pKa = 11.84TRR29 pKa = 11.84AASPARR35 pKa = 11.84IPGIRR40 pKa = 11.84AGSRR44 pKa = 11.84RR45 pKa = 11.84SAKK48 pKa = 9.97LRR50 pKa = 11.84STAATAASRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84ARR63 pKa = 11.84SASTLAASPAGRR75 pKa = 11.84AGSGVPGQAGDD86 pKa = 3.79QRR88 pKa = 11.84PHH90 pKa = 7.18SSASPRR96 pKa = 11.84PQARR100 pKa = 11.84PAITEE105 pKa = 3.91PNGRR109 pKa = 11.84SSTRR113 pKa = 11.84RR114 pKa = 11.84VSSSS118 pKa = 2.59
MM1 pKa = 7.2PRR3 pKa = 11.84EE4 pKa = 3.92RR5 pKa = 11.84QRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84ATVCVVSVSSQAMTSASRR27 pKa = 11.84TRR29 pKa = 11.84AASPARR35 pKa = 11.84IPGIRR40 pKa = 11.84AGSRR44 pKa = 11.84RR45 pKa = 11.84SAKK48 pKa = 9.97LRR50 pKa = 11.84STAATAASRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84ARR63 pKa = 11.84SASTLAASPAGRR75 pKa = 11.84AGSGVPGQAGDD86 pKa = 3.79QRR88 pKa = 11.84PHH90 pKa = 7.18SSASPRR96 pKa = 11.84PQARR100 pKa = 11.84PAITEE105 pKa = 3.91PNGRR109 pKa = 11.84SSTRR113 pKa = 11.84RR114 pKa = 11.84VSSSS118 pKa = 2.59
Molecular weight: 12.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2248913 |
26 |
3549 |
319.2 |
34.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.76 ± 0.043 | 0.773 ± 0.009 |
5.881 ± 0.029 | 5.126 ± 0.026 |
3.463 ± 0.018 | 9.411 ± 0.041 |
1.943 ± 0.014 | 4.729 ± 0.021 |
2.339 ± 0.023 | 10.662 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.141 ± 0.013 | 2.067 ± 0.018 |
5.791 ± 0.023 | 3.044 ± 0.016 |
7.755 ± 0.037 | 4.769 ± 0.021 |
5.181 ± 0.022 | 7.601 ± 0.021 |
1.456 ± 0.013 | 2.103 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
