
Apple-associated luteovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Luteovirus
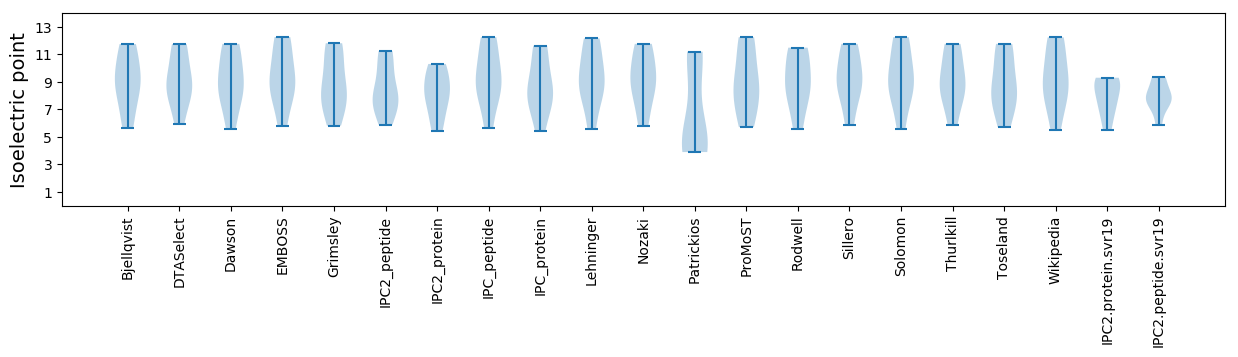
Average proteome isoelectric point is 7.97
Get precalculated fractions of proteins
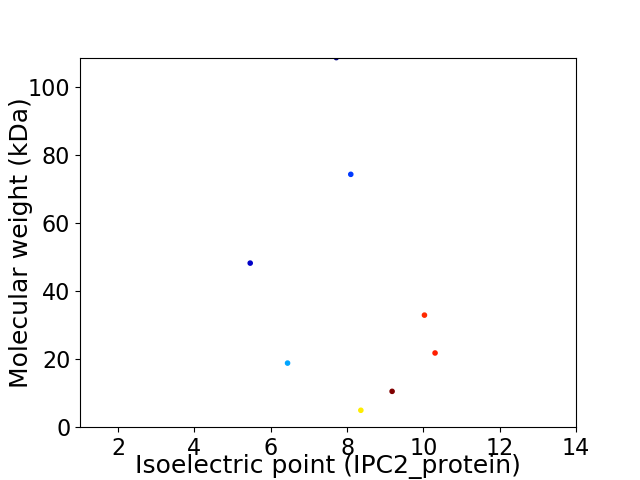
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4QXI9|A0A2H4QXI9_9LUTE Isoform of A0A2H4QXF1 Coat protein OS=Apple-associated luteovirus OX=2054409 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.37FFDD4 pKa = 4.05EE5 pKa = 6.32LITASIKK12 pKa = 9.51VVRR15 pKa = 11.84DD16 pKa = 5.1FIAYY20 pKa = 9.48IYY22 pKa = 11.1NNLKK26 pKa = 10.24NVYY29 pKa = 9.56KK30 pKa = 10.46RR31 pKa = 11.84FKK33 pKa = 8.11MWLWEE38 pKa = 4.01LQGKK42 pKa = 9.06FSQHH46 pKa = 6.33DD47 pKa = 3.63AFADD51 pKa = 3.42MCYY54 pKa = 10.83GYY56 pKa = 9.68MDD58 pKa = 4.34DD59 pKa = 4.04VEE61 pKa = 4.22EE62 pKa = 5.18FEE64 pKa = 5.77WEE66 pKa = 4.3LQTQLDD72 pKa = 3.91NAEE75 pKa = 4.26TQMLLAEE82 pKa = 4.39KK83 pKa = 10.03HH84 pKa = 5.27LQAMLKK90 pKa = 10.14APKK93 pKa = 8.36ITGWPTPTRR102 pKa = 11.84PAGAIPAVPVKK113 pKa = 10.09TRR115 pKa = 11.84TLRR118 pKa = 11.84EE119 pKa = 3.7LADD122 pKa = 3.65EE123 pKa = 4.2VRR125 pKa = 11.84ARR127 pKa = 11.84QGIDD131 pKa = 2.91TGGHH135 pKa = 5.09ATMDD139 pKa = 3.81VPSPSAIPLPDD150 pKa = 4.6LSDD153 pKa = 3.29EE154 pKa = 4.29DD155 pKa = 4.24LRR157 pKa = 11.84FILSDD162 pKa = 3.7TIEE165 pKa = 4.4TDD167 pKa = 3.33PEE169 pKa = 4.33HH170 pKa = 7.39VVVFPPGYY178 pKa = 8.94EE179 pKa = 3.93EE180 pKa = 4.07KK181 pKa = 10.44EE182 pKa = 4.06PRR184 pKa = 11.84VLEE187 pKa = 4.46LPSSPKK193 pKa = 9.91EE194 pKa = 4.01HH195 pKa = 7.28IIQEE199 pKa = 4.56PIATLGQAYY208 pKa = 8.11TPEE211 pKa = 6.11DD212 pKa = 3.43IEE214 pKa = 4.11KK215 pKa = 10.76ARR217 pKa = 11.84AEE219 pKa = 4.14FARR222 pKa = 11.84ARR224 pKa = 11.84LDD226 pKa = 3.36YY227 pKa = 9.47TVCLEE232 pKa = 4.21EE233 pKa = 4.27TLDD236 pKa = 3.72AYY238 pKa = 10.06EE239 pKa = 4.39AEE241 pKa = 4.57KK242 pKa = 10.99GDD244 pKa = 3.93GYY246 pKa = 10.91FGRR249 pKa = 11.84FFNTVAHH256 pKa = 6.32RR257 pKa = 11.84MAYY260 pKa = 9.21IKK262 pKa = 10.31KK263 pKa = 9.46CKK265 pKa = 9.77ARR267 pKa = 11.84RR268 pKa = 11.84AKK270 pKa = 9.95TDD272 pKa = 3.67LLAHH276 pKa = 6.99KK277 pKa = 9.79ISNRR281 pKa = 11.84VRR283 pKa = 11.84SAPSVAEE290 pKa = 3.77FHH292 pKa = 6.55SLCNVVEE299 pKa = 4.37EE300 pKa = 4.45PTGEE304 pKa = 4.01YY305 pKa = 10.84KK306 pKa = 10.69NIHH309 pKa = 6.25KK310 pKa = 10.74DD311 pKa = 2.85EE312 pKa = 4.59GEE314 pKa = 4.0NLRR317 pKa = 11.84EE318 pKa = 3.87EE319 pKa = 4.33VMKK322 pKa = 10.03VVRR325 pKa = 11.84YY326 pKa = 9.58IKK328 pKa = 9.57PDD330 pKa = 3.25KK331 pKa = 10.84VRR333 pKa = 11.84EE334 pKa = 3.85AQQYY338 pKa = 7.5IRR340 pKa = 11.84HH341 pKa = 5.08YY342 pKa = 11.12VRR344 pKa = 11.84AKK346 pKa = 10.51NNRR349 pKa = 11.84LSADD353 pKa = 3.73EE354 pKa = 4.35VSSATINRR362 pKa = 11.84YY363 pKa = 6.97VQQFAEE369 pKa = 4.41DD370 pKa = 3.77NKK372 pKa = 10.92LSLSSTHH379 pKa = 6.24LLIRR383 pKa = 11.84AALTMVPVITKK394 pKa = 9.8EE395 pKa = 3.98DD396 pKa = 3.61MMTAMVIHH404 pKa = 6.66GPAARR409 pKa = 11.84KK410 pKa = 9.28ARR412 pKa = 11.84SDD414 pKa = 3.39LSTLEE419 pKa = 4.5GGDD422 pKa = 3.64FF423 pKa = 3.8
MM1 pKa = 7.37FFDD4 pKa = 4.05EE5 pKa = 6.32LITASIKK12 pKa = 9.51VVRR15 pKa = 11.84DD16 pKa = 5.1FIAYY20 pKa = 9.48IYY22 pKa = 11.1NNLKK26 pKa = 10.24NVYY29 pKa = 9.56KK30 pKa = 10.46RR31 pKa = 11.84FKK33 pKa = 8.11MWLWEE38 pKa = 4.01LQGKK42 pKa = 9.06FSQHH46 pKa = 6.33DD47 pKa = 3.63AFADD51 pKa = 3.42MCYY54 pKa = 10.83GYY56 pKa = 9.68MDD58 pKa = 4.34DD59 pKa = 4.04VEE61 pKa = 4.22EE62 pKa = 5.18FEE64 pKa = 5.77WEE66 pKa = 4.3LQTQLDD72 pKa = 3.91NAEE75 pKa = 4.26TQMLLAEE82 pKa = 4.39KK83 pKa = 10.03HH84 pKa = 5.27LQAMLKK90 pKa = 10.14APKK93 pKa = 8.36ITGWPTPTRR102 pKa = 11.84PAGAIPAVPVKK113 pKa = 10.09TRR115 pKa = 11.84TLRR118 pKa = 11.84EE119 pKa = 3.7LADD122 pKa = 3.65EE123 pKa = 4.2VRR125 pKa = 11.84ARR127 pKa = 11.84QGIDD131 pKa = 2.91TGGHH135 pKa = 5.09ATMDD139 pKa = 3.81VPSPSAIPLPDD150 pKa = 4.6LSDD153 pKa = 3.29EE154 pKa = 4.29DD155 pKa = 4.24LRR157 pKa = 11.84FILSDD162 pKa = 3.7TIEE165 pKa = 4.4TDD167 pKa = 3.33PEE169 pKa = 4.33HH170 pKa = 7.39VVVFPPGYY178 pKa = 8.94EE179 pKa = 3.93EE180 pKa = 4.07KK181 pKa = 10.44EE182 pKa = 4.06PRR184 pKa = 11.84VLEE187 pKa = 4.46LPSSPKK193 pKa = 9.91EE194 pKa = 4.01HH195 pKa = 7.28IIQEE199 pKa = 4.56PIATLGQAYY208 pKa = 8.11TPEE211 pKa = 6.11DD212 pKa = 3.43IEE214 pKa = 4.11KK215 pKa = 10.76ARR217 pKa = 11.84AEE219 pKa = 4.14FARR222 pKa = 11.84ARR224 pKa = 11.84LDD226 pKa = 3.36YY227 pKa = 9.47TVCLEE232 pKa = 4.21EE233 pKa = 4.27TLDD236 pKa = 3.72AYY238 pKa = 10.06EE239 pKa = 4.39AEE241 pKa = 4.57KK242 pKa = 10.99GDD244 pKa = 3.93GYY246 pKa = 10.91FGRR249 pKa = 11.84FFNTVAHH256 pKa = 6.32RR257 pKa = 11.84MAYY260 pKa = 9.21IKK262 pKa = 10.31KK263 pKa = 9.46CKK265 pKa = 9.77ARR267 pKa = 11.84RR268 pKa = 11.84AKK270 pKa = 9.95TDD272 pKa = 3.67LLAHH276 pKa = 6.99KK277 pKa = 9.79ISNRR281 pKa = 11.84VRR283 pKa = 11.84SAPSVAEE290 pKa = 3.77FHH292 pKa = 6.55SLCNVVEE299 pKa = 4.37EE300 pKa = 4.45PTGEE304 pKa = 4.01YY305 pKa = 10.84KK306 pKa = 10.69NIHH309 pKa = 6.25KK310 pKa = 10.74DD311 pKa = 2.85EE312 pKa = 4.59GEE314 pKa = 4.0NLRR317 pKa = 11.84EE318 pKa = 3.87EE319 pKa = 4.33VMKK322 pKa = 10.03VVRR325 pKa = 11.84YY326 pKa = 9.58IKK328 pKa = 9.57PDD330 pKa = 3.25KK331 pKa = 10.84VRR333 pKa = 11.84EE334 pKa = 3.85AQQYY338 pKa = 7.5IRR340 pKa = 11.84HH341 pKa = 5.08YY342 pKa = 11.12VRR344 pKa = 11.84AKK346 pKa = 10.51NNRR349 pKa = 11.84LSADD353 pKa = 3.73EE354 pKa = 4.35VSSATINRR362 pKa = 11.84YY363 pKa = 6.97VQQFAEE369 pKa = 4.41DD370 pKa = 3.77NKK372 pKa = 10.92LSLSSTHH379 pKa = 6.24LLIRR383 pKa = 11.84AALTMVPVITKK394 pKa = 9.8EE395 pKa = 3.98DD396 pKa = 3.61MMTAMVIHH404 pKa = 6.66GPAARR409 pKa = 11.84KK410 pKa = 9.28ARR412 pKa = 11.84SDD414 pKa = 3.39LSTLEE419 pKa = 4.5GGDD422 pKa = 3.64FF423 pKa = 3.8
Molecular weight: 48.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4QXD9|A0A2H4QXD9_9LUTE Movement protein OS=Apple-associated luteovirus OX=2054409 GN=ORF4 PE=3 SV=1
MM1 pKa = 7.63RR2 pKa = 11.84AVDD5 pKa = 3.44SWRR8 pKa = 11.84VTHH11 pKa = 6.19VKK13 pKa = 10.43NMPPPRR19 pKa = 11.84GVDD22 pKa = 3.43CLGLRR27 pKa = 11.84HH28 pKa = 6.95RR29 pKa = 11.84ITSVHH34 pKa = 5.54VLGLSEE40 pKa = 4.22VLWVVEE46 pKa = 4.28ACLHH50 pKa = 5.64IHH52 pKa = 4.73TCRR55 pKa = 11.84WKK57 pKa = 9.75GSGGPVHH64 pKa = 6.35TLHH67 pKa = 6.37GVVRR71 pKa = 11.84IPKK74 pKa = 8.42RR75 pKa = 11.84TSSDD79 pKa = 3.48SLTMNQSTDD88 pKa = 2.83CGYY91 pKa = 11.06SRR93 pKa = 11.84RR94 pKa = 3.99
MM1 pKa = 7.63RR2 pKa = 11.84AVDD5 pKa = 3.44SWRR8 pKa = 11.84VTHH11 pKa = 6.19VKK13 pKa = 10.43NMPPPRR19 pKa = 11.84GVDD22 pKa = 3.43CLGLRR27 pKa = 11.84HH28 pKa = 6.95RR29 pKa = 11.84ITSVHH34 pKa = 5.54VLGLSEE40 pKa = 4.22VLWVVEE46 pKa = 4.28ACLHH50 pKa = 5.64IHH52 pKa = 4.73TCRR55 pKa = 11.84WKK57 pKa = 9.75GSGGPVHH64 pKa = 6.35TLHH67 pKa = 6.37GVVRR71 pKa = 11.84IPKK74 pKa = 8.42RR75 pKa = 11.84TSSDD79 pKa = 3.48SLTMNQSTDD88 pKa = 2.83CGYY91 pKa = 11.06SRR93 pKa = 11.84RR94 pKa = 3.99
Molecular weight: 10.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2851 |
45 |
951 |
356.4 |
40.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.155 ± 0.808 | 1.684 ± 0.466 |
5.367 ± 0.935 | 5.542 ± 1.124 |
3.718 ± 0.387 | 5.928 ± 0.424 |
2.21 ± 0.415 | 4.981 ± 0.51 |
5.437 ± 0.592 | 8.909 ± 0.748 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.463 | 3.578 ± 0.387 |
5.893 ± 0.547 | 3.297 ± 0.17 |
8.313 ± 0.886 | 8.839 ± 1.584 |
5.787 ± 0.762 | 6.559 ± 0.662 |
1.263 ± 0.18 | 2.911 ± 0.385 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
