
Streptococcus satellite phage Javan225
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
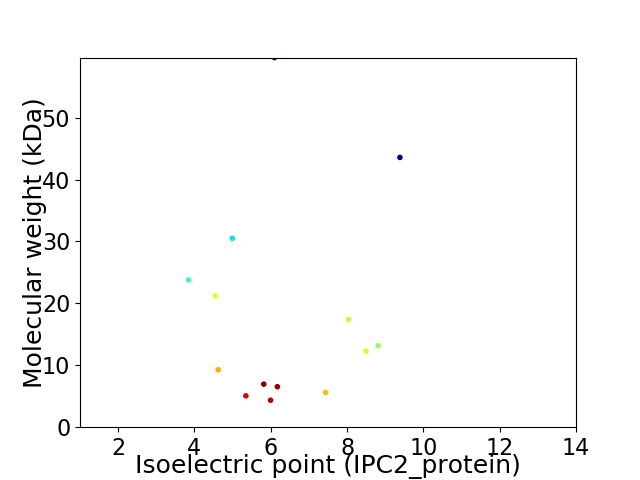
Virtual 2D-PAGE plot for 14 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZHR0|A0A4D5ZHR0_9VIRU Integrase/recombinase OS=Streptococcus satellite phage Javan225 OX=2558580 GN=JavanS225_0001 PE=3 SV=1
MM1 pKa = 7.1KK2 pKa = 10.41HH3 pKa = 6.2EE4 pKa = 4.54EE5 pKa = 4.04LTLTEE10 pKa = 4.31LKK12 pKa = 10.64DD13 pKa = 3.32WTKK16 pKa = 10.5SARR19 pKa = 11.84NLVEE23 pKa = 4.73HH24 pKa = 6.5LQDD27 pKa = 5.13DD28 pKa = 4.2YY29 pKa = 11.95HH30 pKa = 7.94DD31 pKa = 4.23YY32 pKa = 11.44LPTGKK37 pKa = 10.28AEE39 pKa = 4.26AFGSITNSIYY49 pKa = 11.19NCLDD53 pKa = 3.3CLAEE57 pKa = 4.17MFDD60 pKa = 3.73NDD62 pKa = 3.49EE63 pKa = 4.65LEE65 pKa = 4.24LVEE68 pKa = 4.15YY69 pKa = 9.87TIDD72 pKa = 3.83PQEE75 pKa = 3.93TTIDD79 pKa = 3.77DD80 pKa = 3.9NQADD84 pKa = 4.66EE85 pKa = 4.0IAQYY89 pKa = 10.89NEE91 pKa = 3.57LMKK94 pKa = 10.63EE95 pKa = 4.13VEE97 pKa = 4.64DD98 pKa = 3.69SDD100 pKa = 4.11KK101 pKa = 10.52EE102 pKa = 3.93ARR104 pKa = 11.84EE105 pKa = 4.15IVCYY109 pKa = 10.15EE110 pKa = 3.89YY111 pKa = 11.07DD112 pKa = 3.84DD113 pKa = 5.08TIHH116 pKa = 6.98EE117 pKa = 4.72YY118 pKa = 9.73IDD120 pKa = 3.8KK121 pKa = 9.55RR122 pKa = 11.84TEE124 pKa = 3.8QLKK127 pKa = 9.5EE128 pKa = 3.5QATFEE133 pKa = 4.0EE134 pKa = 4.62LAYY137 pKa = 9.82KK138 pKa = 9.5VALYY142 pKa = 10.32EE143 pKa = 5.07SEE145 pKa = 4.11LLDD148 pKa = 3.57YY149 pKa = 11.28AEE151 pKa = 5.23RR152 pKa = 11.84LLSDD156 pKa = 3.74EE157 pKa = 4.79PLNADD162 pKa = 3.49SEE164 pKa = 4.84TAIGTLDD171 pKa = 3.63MLDD174 pKa = 4.84DD175 pKa = 3.92EE176 pKa = 6.32AIDD179 pKa = 3.89LFKK182 pKa = 11.28SVDD185 pKa = 3.22VDD187 pKa = 3.55NEE189 pKa = 4.36YY190 pKa = 11.0QGLEE194 pKa = 4.14YY195 pKa = 11.23YY196 pKa = 8.19NTSLNKK202 pKa = 9.95EE203 pKa = 4.21DD204 pKa = 3.57
MM1 pKa = 7.1KK2 pKa = 10.41HH3 pKa = 6.2EE4 pKa = 4.54EE5 pKa = 4.04LTLTEE10 pKa = 4.31LKK12 pKa = 10.64DD13 pKa = 3.32WTKK16 pKa = 10.5SARR19 pKa = 11.84NLVEE23 pKa = 4.73HH24 pKa = 6.5LQDD27 pKa = 5.13DD28 pKa = 4.2YY29 pKa = 11.95HH30 pKa = 7.94DD31 pKa = 4.23YY32 pKa = 11.44LPTGKK37 pKa = 10.28AEE39 pKa = 4.26AFGSITNSIYY49 pKa = 11.19NCLDD53 pKa = 3.3CLAEE57 pKa = 4.17MFDD60 pKa = 3.73NDD62 pKa = 3.49EE63 pKa = 4.65LEE65 pKa = 4.24LVEE68 pKa = 4.15YY69 pKa = 9.87TIDD72 pKa = 3.83PQEE75 pKa = 3.93TTIDD79 pKa = 3.77DD80 pKa = 3.9NQADD84 pKa = 4.66EE85 pKa = 4.0IAQYY89 pKa = 10.89NEE91 pKa = 3.57LMKK94 pKa = 10.63EE95 pKa = 4.13VEE97 pKa = 4.64DD98 pKa = 3.69SDD100 pKa = 4.11KK101 pKa = 10.52EE102 pKa = 3.93ARR104 pKa = 11.84EE105 pKa = 4.15IVCYY109 pKa = 10.15EE110 pKa = 3.89YY111 pKa = 11.07DD112 pKa = 3.84DD113 pKa = 5.08TIHH116 pKa = 6.98EE117 pKa = 4.72YY118 pKa = 9.73IDD120 pKa = 3.8KK121 pKa = 9.55RR122 pKa = 11.84TEE124 pKa = 3.8QLKK127 pKa = 9.5EE128 pKa = 3.5QATFEE133 pKa = 4.0EE134 pKa = 4.62LAYY137 pKa = 9.82KK138 pKa = 9.5VALYY142 pKa = 10.32EE143 pKa = 5.07SEE145 pKa = 4.11LLDD148 pKa = 3.57YY149 pKa = 11.28AEE151 pKa = 5.23RR152 pKa = 11.84LLSDD156 pKa = 3.74EE157 pKa = 4.79PLNADD162 pKa = 3.49SEE164 pKa = 4.84TAIGTLDD171 pKa = 3.63MLDD174 pKa = 4.84DD175 pKa = 3.92EE176 pKa = 6.32AIDD179 pKa = 3.89LFKK182 pKa = 11.28SVDD185 pKa = 3.22VDD187 pKa = 3.55NEE189 pKa = 4.36YY190 pKa = 11.0QGLEE194 pKa = 4.14YY195 pKa = 11.23YY196 pKa = 8.19NTSLNKK202 pKa = 9.95EE203 pKa = 4.21DD204 pKa = 3.57
Molecular weight: 23.78 kDa
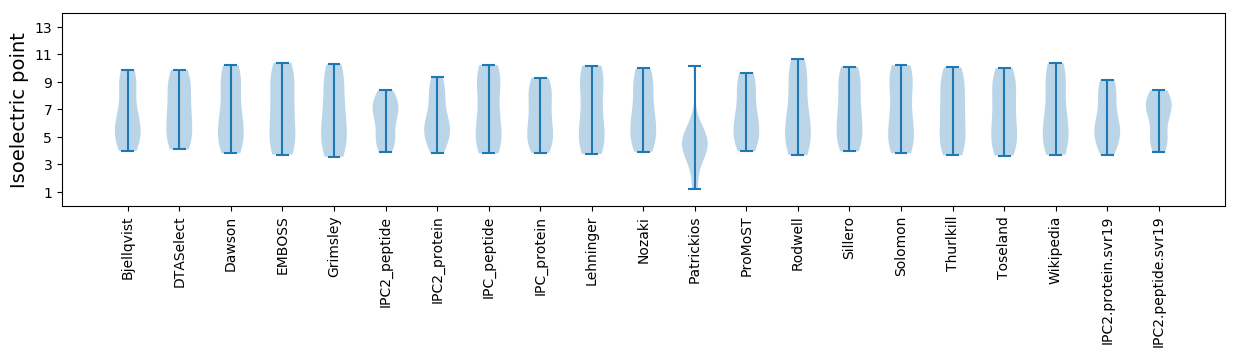
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZHS1|A0A4D5ZHS1_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan225 OX=2558580 GN=JavanS225_0011 PE=4 SV=1
MM1 pKa = 7.51NIKK4 pKa = 10.04EE5 pKa = 4.29VIKK8 pKa = 10.83KK9 pKa = 10.43DD10 pKa = 3.21GTKK13 pKa = 9.85VYY15 pKa = 10.42RR16 pKa = 11.84SNVYY20 pKa = 10.53LGVDD24 pKa = 4.04SITGKK29 pKa = 10.18KK30 pKa = 10.14VKK32 pKa = 8.5TTVTGRR38 pKa = 11.84TKK40 pKa = 10.89KK41 pKa = 9.66EE42 pKa = 4.03VKK44 pKa = 9.46TKK46 pKa = 10.56AQQAQSNFKK55 pKa = 11.0ANGSTVFKK63 pKa = 10.15RR64 pKa = 11.84VEE66 pKa = 3.73VTTYY70 pKa = 11.13KK71 pKa = 10.76EE72 pKa = 4.39LTDD75 pKa = 3.82LWLEE79 pKa = 4.13NYY81 pKa = 10.11QMTVKK86 pKa = 10.31PQTLVNTRR94 pKa = 11.84QFLRR98 pKa = 11.84NHH100 pKa = 6.87ILPVFGDD107 pKa = 3.56MQLDD111 pKa = 4.24KK112 pKa = 11.2IHH114 pKa = 6.9IAHH117 pKa = 6.2IQSWVNKK124 pKa = 9.54LAFKK128 pKa = 10.19IVNYY132 pKa = 10.05GVAASINKK140 pKa = 9.97RR141 pKa = 11.84ILQYY145 pKa = 10.53GVSMQLIPFNPARR158 pKa = 11.84EE159 pKa = 4.31VILPRR164 pKa = 11.84PQKK167 pKa = 10.82AGANRR172 pKa = 11.84IKK174 pKa = 10.79FIDD177 pKa = 3.91KK178 pKa = 10.6EE179 pKa = 4.13DD180 pKa = 3.62LKK182 pKa = 10.6TFLDD186 pKa = 3.69YY187 pKa = 10.59MEE189 pKa = 5.45RR190 pKa = 11.84LAPTAYY196 pKa = 10.16NYY198 pKa = 11.09YY199 pKa = 9.71YY200 pKa = 11.02DD201 pKa = 3.73SVLYY205 pKa = 10.49KK206 pKa = 10.87LLLATGCRR214 pKa = 11.84YY215 pKa = 10.48GEE217 pKa = 4.05AVALEE222 pKa = 4.14WSDD225 pKa = 3.78IDD227 pKa = 5.0FDD229 pKa = 3.99NATINITKK237 pKa = 9.12TYY239 pKa = 9.96NRR241 pKa = 11.84IVKK244 pKa = 9.79QVGTPKK250 pKa = 10.27SKK252 pKa = 10.81AGIRR256 pKa = 11.84IISIDD261 pKa = 3.43NKK263 pKa = 9.44TILMLKK269 pKa = 9.54QYY271 pKa = 10.83RR272 pKa = 11.84NRR274 pKa = 11.84QRR276 pKa = 11.84QAFMEE281 pKa = 4.42IGAPAPALVFSTTISQYY298 pKa = 9.95PNSDD302 pKa = 2.89ARR304 pKa = 11.84TKK306 pKa = 10.38SLRR309 pKa = 11.84HH310 pKa = 5.31RR311 pKa = 11.84CKK313 pKa = 10.41EE314 pKa = 3.57AGIPQFTFHH323 pKa = 7.42AFRR326 pKa = 11.84HH327 pKa = 4.69THH329 pKa = 6.92ASLLLNAGIGYY340 pKa = 9.74KK341 pKa = 9.96EE342 pKa = 3.9LQHH345 pKa = 6.67RR346 pKa = 11.84LGHH349 pKa = 6.08ATLAMTMDD357 pKa = 4.61TYY359 pKa = 11.85SHH361 pKa = 7.05LSKK364 pKa = 10.62EE365 pKa = 4.29KK366 pKa = 9.9EE367 pKa = 4.1KK368 pKa = 11.05EE369 pKa = 3.73AVLYY373 pKa = 10.0YY374 pKa = 10.87GKK376 pKa = 10.6ALQNLL381 pKa = 3.91
MM1 pKa = 7.51NIKK4 pKa = 10.04EE5 pKa = 4.29VIKK8 pKa = 10.83KK9 pKa = 10.43DD10 pKa = 3.21GTKK13 pKa = 9.85VYY15 pKa = 10.42RR16 pKa = 11.84SNVYY20 pKa = 10.53LGVDD24 pKa = 4.04SITGKK29 pKa = 10.18KK30 pKa = 10.14VKK32 pKa = 8.5TTVTGRR38 pKa = 11.84TKK40 pKa = 10.89KK41 pKa = 9.66EE42 pKa = 4.03VKK44 pKa = 9.46TKK46 pKa = 10.56AQQAQSNFKK55 pKa = 11.0ANGSTVFKK63 pKa = 10.15RR64 pKa = 11.84VEE66 pKa = 3.73VTTYY70 pKa = 11.13KK71 pKa = 10.76EE72 pKa = 4.39LTDD75 pKa = 3.82LWLEE79 pKa = 4.13NYY81 pKa = 10.11QMTVKK86 pKa = 10.31PQTLVNTRR94 pKa = 11.84QFLRR98 pKa = 11.84NHH100 pKa = 6.87ILPVFGDD107 pKa = 3.56MQLDD111 pKa = 4.24KK112 pKa = 11.2IHH114 pKa = 6.9IAHH117 pKa = 6.2IQSWVNKK124 pKa = 9.54LAFKK128 pKa = 10.19IVNYY132 pKa = 10.05GVAASINKK140 pKa = 9.97RR141 pKa = 11.84ILQYY145 pKa = 10.53GVSMQLIPFNPARR158 pKa = 11.84EE159 pKa = 4.31VILPRR164 pKa = 11.84PQKK167 pKa = 10.82AGANRR172 pKa = 11.84IKK174 pKa = 10.79FIDD177 pKa = 3.91KK178 pKa = 10.6EE179 pKa = 4.13DD180 pKa = 3.62LKK182 pKa = 10.6TFLDD186 pKa = 3.69YY187 pKa = 10.59MEE189 pKa = 5.45RR190 pKa = 11.84LAPTAYY196 pKa = 10.16NYY198 pKa = 11.09YY199 pKa = 9.71YY200 pKa = 11.02DD201 pKa = 3.73SVLYY205 pKa = 10.49KK206 pKa = 10.87LLLATGCRR214 pKa = 11.84YY215 pKa = 10.48GEE217 pKa = 4.05AVALEE222 pKa = 4.14WSDD225 pKa = 3.78IDD227 pKa = 5.0FDD229 pKa = 3.99NATINITKK237 pKa = 9.12TYY239 pKa = 9.96NRR241 pKa = 11.84IVKK244 pKa = 9.79QVGTPKK250 pKa = 10.27SKK252 pKa = 10.81AGIRR256 pKa = 11.84IISIDD261 pKa = 3.43NKK263 pKa = 9.44TILMLKK269 pKa = 9.54QYY271 pKa = 10.83RR272 pKa = 11.84NRR274 pKa = 11.84QRR276 pKa = 11.84QAFMEE281 pKa = 4.42IGAPAPALVFSTTISQYY298 pKa = 9.95PNSDD302 pKa = 2.89ARR304 pKa = 11.84TKK306 pKa = 10.38SLRR309 pKa = 11.84HH310 pKa = 5.31RR311 pKa = 11.84CKK313 pKa = 10.41EE314 pKa = 3.57AGIPQFTFHH323 pKa = 7.42AFRR326 pKa = 11.84HH327 pKa = 4.69THH329 pKa = 6.92ASLLLNAGIGYY340 pKa = 9.74KK341 pKa = 9.96EE342 pKa = 3.9LQHH345 pKa = 6.67RR346 pKa = 11.84LGHH349 pKa = 6.08ATLAMTMDD357 pKa = 4.61TYY359 pKa = 11.85SHH361 pKa = 7.05LSKK364 pKa = 10.62EE365 pKa = 4.29KK366 pKa = 9.9EE367 pKa = 4.1KK368 pKa = 11.05EE369 pKa = 3.73AVLYY373 pKa = 10.0YY374 pKa = 10.87GKK376 pKa = 10.6ALQNLL381 pKa = 3.91
Molecular weight: 43.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2236 |
37 |
517 |
159.7 |
18.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.68 ± 0.818 | 0.85 ± 0.151 |
6.172 ± 0.79 | 8.274 ± 0.994 |
4.562 ± 0.458 | 4.517 ± 0.557 |
1.565 ± 0.229 | 7.245 ± 0.431 |
8.855 ± 0.677 | 10.018 ± 0.689 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.549 ± 0.299 | 6.574 ± 0.51 |
3.354 ± 0.496 | 4.204 ± 0.44 |
4.07 ± 0.514 | 4.293 ± 0.327 |
5.814 ± 0.473 | 5.367 ± 0.431 |
1.029 ± 0.219 | 5.009 ± 0.404 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
