
Little cherry virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Velarivirus
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
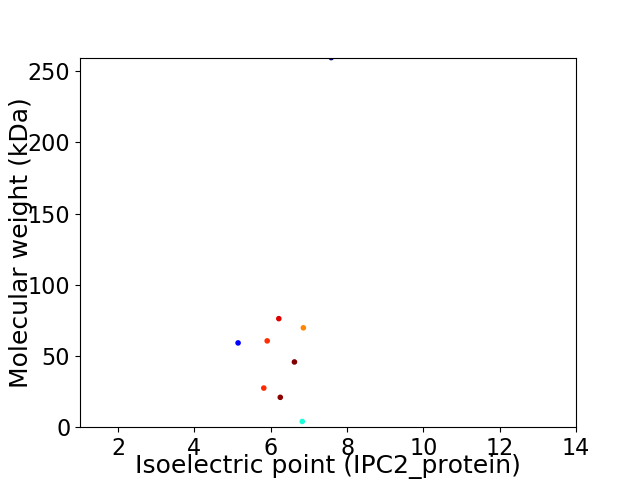
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
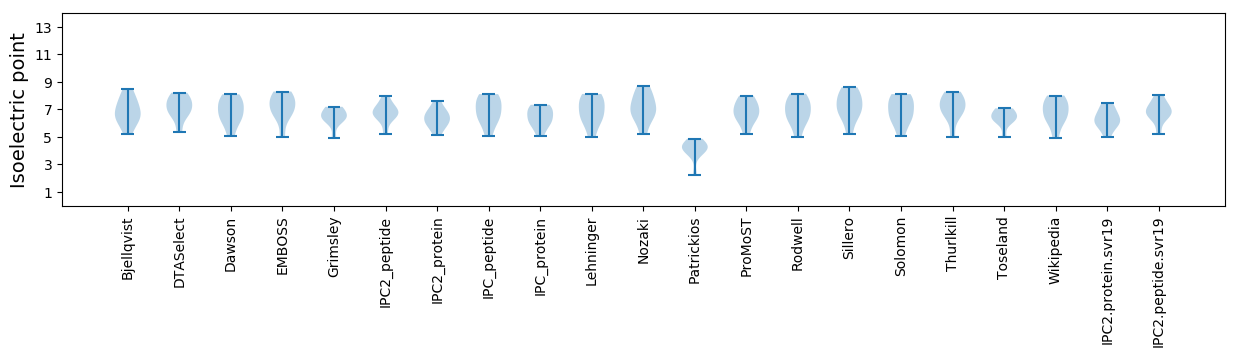
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O40961|O40961_9CLOS Uncharacterized protein OS=Little cherry virus 1 OX=217686 PE=4 SV=1
MM1 pKa = 7.86PNLGSDD7 pKa = 3.81SVFDD11 pKa = 4.18EE12 pKa = 4.59YY13 pKa = 11.7SLVMEE18 pKa = 5.01DD19 pKa = 3.43RR20 pKa = 11.84PFNVFPARR28 pKa = 11.84PSLSSVHH35 pKa = 6.07AVNDD39 pKa = 4.0FMDD42 pKa = 3.66LTFNGLSSVDD52 pKa = 3.41YY53 pKa = 9.02LTRR56 pKa = 11.84TLRR59 pKa = 11.84FEE61 pKa = 4.27YY62 pKa = 10.72SDD64 pKa = 3.75IEE66 pKa = 4.4LPCLEE71 pKa = 5.39DD72 pKa = 5.32LLLTPTKK79 pKa = 10.24SKK81 pKa = 10.32PYY83 pKa = 9.73QAVNCKK89 pKa = 10.06VPILIGKK96 pKa = 7.52GEE98 pKa = 4.27RR99 pKa = 11.84SRR101 pKa = 11.84PDD103 pKa = 2.47TWKK106 pKa = 10.59QVILSLSKK114 pKa = 10.92RR115 pKa = 11.84NFAAPRR121 pKa = 11.84LNEE124 pKa = 5.19DD125 pKa = 2.83ISIDD129 pKa = 3.47EE130 pKa = 4.22TANRR134 pKa = 11.84LLTSLMRR141 pKa = 11.84CMEE144 pKa = 4.66PSRR147 pKa = 11.84LAEE150 pKa = 4.0YY151 pKa = 10.5FDD153 pKa = 4.15VVEE156 pKa = 4.49PDD158 pKa = 3.36VNKK161 pKa = 10.36INSWLLTRR169 pKa = 11.84DD170 pKa = 2.84RR171 pKa = 11.84RR172 pKa = 11.84KK173 pKa = 10.2YY174 pKa = 10.57GNVLRR179 pKa = 11.84GFDD182 pKa = 3.84GNDD185 pKa = 2.67WTTKK189 pKa = 10.44LFNLKK194 pKa = 10.57LMVKK198 pKa = 10.58GDD200 pKa = 4.33MKK202 pKa = 10.8PKK204 pKa = 10.33LDD206 pKa = 3.44MSGVSEE212 pKa = 4.38YY213 pKa = 11.24SPASNIIYY221 pKa = 8.44YY222 pKa = 8.24QTMINMFFSPIFLEE236 pKa = 3.56IFGRR240 pKa = 11.84IKK242 pKa = 10.71YY243 pKa = 10.17CLGDD247 pKa = 3.66KK248 pKa = 10.72VILFSGMNLDD258 pKa = 3.73EE259 pKa = 4.97MGDD262 pKa = 3.7LLEE265 pKa = 5.72SRR267 pKa = 11.84LCYY270 pKa = 9.83PLNSYY275 pKa = 11.06NFVEE279 pKa = 4.21VDD281 pKa = 3.99FSKK284 pKa = 10.7FDD286 pKa = 3.23KK287 pKa = 11.13SQGHH291 pKa = 5.4VIKK294 pKa = 10.59LYY296 pKa = 10.74EE297 pKa = 4.0EE298 pKa = 4.01LVYY301 pKa = 10.67KK302 pKa = 10.54IFKK305 pKa = 10.39FSPNLYY311 pKa = 10.7DD312 pKa = 3.53NFKK315 pKa = 10.58LSEE318 pKa = 4.22YY319 pKa = 10.0FCRR322 pKa = 11.84ASATCGVNLDD332 pKa = 4.88LYY334 pKa = 8.75CQRR337 pKa = 11.84RR338 pKa = 11.84TGSPNTWLSNSLATLAMLASVYY360 pKa = 10.95DD361 pKa = 4.34LDD363 pKa = 7.01DD364 pKa = 4.14IDD366 pKa = 6.11LIIVSGDD373 pKa = 3.18DD374 pKa = 3.23SLIISRR380 pKa = 11.84KK381 pKa = 9.13EE382 pKa = 3.63IEE384 pKa = 4.42NKK386 pKa = 10.21CLEE389 pKa = 4.18INNDD393 pKa = 3.05FGMDD397 pKa = 3.38AKK399 pKa = 10.76FLANPVPYY407 pKa = 9.7FCSKK411 pKa = 10.55FIIQVDD417 pKa = 3.27NRR419 pKa = 11.84IRR421 pKa = 11.84LVPDD425 pKa = 3.63PVRR428 pKa = 11.84FFEE431 pKa = 4.54KK432 pKa = 10.51LSTPVTLVQLEE443 pKa = 4.56HH444 pKa = 6.44PTLLRR449 pKa = 11.84EE450 pKa = 4.11RR451 pKa = 11.84FTSYY455 pKa = 10.73RR456 pKa = 11.84DD457 pKa = 3.36LMGSYY462 pKa = 10.25FDD464 pKa = 4.18EE465 pKa = 4.15NVIIAVEE472 pKa = 4.08RR473 pKa = 11.84FVSLKK478 pKa = 10.38YY479 pKa = 8.06NTPMGSGYY487 pKa = 11.01AAFCYY492 pKa = 9.66IHH494 pKa = 7.1CLLSSFKK501 pKa = 11.01NFLTIFGDD509 pKa = 3.65DD510 pKa = 3.41TSIQLL515 pKa = 3.8
MM1 pKa = 7.86PNLGSDD7 pKa = 3.81SVFDD11 pKa = 4.18EE12 pKa = 4.59YY13 pKa = 11.7SLVMEE18 pKa = 5.01DD19 pKa = 3.43RR20 pKa = 11.84PFNVFPARR28 pKa = 11.84PSLSSVHH35 pKa = 6.07AVNDD39 pKa = 4.0FMDD42 pKa = 3.66LTFNGLSSVDD52 pKa = 3.41YY53 pKa = 9.02LTRR56 pKa = 11.84TLRR59 pKa = 11.84FEE61 pKa = 4.27YY62 pKa = 10.72SDD64 pKa = 3.75IEE66 pKa = 4.4LPCLEE71 pKa = 5.39DD72 pKa = 5.32LLLTPTKK79 pKa = 10.24SKK81 pKa = 10.32PYY83 pKa = 9.73QAVNCKK89 pKa = 10.06VPILIGKK96 pKa = 7.52GEE98 pKa = 4.27RR99 pKa = 11.84SRR101 pKa = 11.84PDD103 pKa = 2.47TWKK106 pKa = 10.59QVILSLSKK114 pKa = 10.92RR115 pKa = 11.84NFAAPRR121 pKa = 11.84LNEE124 pKa = 5.19DD125 pKa = 2.83ISIDD129 pKa = 3.47EE130 pKa = 4.22TANRR134 pKa = 11.84LLTSLMRR141 pKa = 11.84CMEE144 pKa = 4.66PSRR147 pKa = 11.84LAEE150 pKa = 4.0YY151 pKa = 10.5FDD153 pKa = 4.15VVEE156 pKa = 4.49PDD158 pKa = 3.36VNKK161 pKa = 10.36INSWLLTRR169 pKa = 11.84DD170 pKa = 2.84RR171 pKa = 11.84RR172 pKa = 11.84KK173 pKa = 10.2YY174 pKa = 10.57GNVLRR179 pKa = 11.84GFDD182 pKa = 3.84GNDD185 pKa = 2.67WTTKK189 pKa = 10.44LFNLKK194 pKa = 10.57LMVKK198 pKa = 10.58GDD200 pKa = 4.33MKK202 pKa = 10.8PKK204 pKa = 10.33LDD206 pKa = 3.44MSGVSEE212 pKa = 4.38YY213 pKa = 11.24SPASNIIYY221 pKa = 8.44YY222 pKa = 8.24QTMINMFFSPIFLEE236 pKa = 3.56IFGRR240 pKa = 11.84IKK242 pKa = 10.71YY243 pKa = 10.17CLGDD247 pKa = 3.66KK248 pKa = 10.72VILFSGMNLDD258 pKa = 3.73EE259 pKa = 4.97MGDD262 pKa = 3.7LLEE265 pKa = 5.72SRR267 pKa = 11.84LCYY270 pKa = 9.83PLNSYY275 pKa = 11.06NFVEE279 pKa = 4.21VDD281 pKa = 3.99FSKK284 pKa = 10.7FDD286 pKa = 3.23KK287 pKa = 11.13SQGHH291 pKa = 5.4VIKK294 pKa = 10.59LYY296 pKa = 10.74EE297 pKa = 4.0EE298 pKa = 4.01LVYY301 pKa = 10.67KK302 pKa = 10.54IFKK305 pKa = 10.39FSPNLYY311 pKa = 10.7DD312 pKa = 3.53NFKK315 pKa = 10.58LSEE318 pKa = 4.22YY319 pKa = 10.0FCRR322 pKa = 11.84ASATCGVNLDD332 pKa = 4.88LYY334 pKa = 8.75CQRR337 pKa = 11.84RR338 pKa = 11.84TGSPNTWLSNSLATLAMLASVYY360 pKa = 10.95DD361 pKa = 4.34LDD363 pKa = 7.01DD364 pKa = 4.14IDD366 pKa = 6.11LIIVSGDD373 pKa = 3.18DD374 pKa = 3.23SLIISRR380 pKa = 11.84KK381 pKa = 9.13EE382 pKa = 3.63IEE384 pKa = 4.42NKK386 pKa = 10.21CLEE389 pKa = 4.18INNDD393 pKa = 3.05FGMDD397 pKa = 3.38AKK399 pKa = 10.76FLANPVPYY407 pKa = 9.7FCSKK411 pKa = 10.55FIIQVDD417 pKa = 3.27NRR419 pKa = 11.84IRR421 pKa = 11.84LVPDD425 pKa = 3.63PVRR428 pKa = 11.84FFEE431 pKa = 4.54KK432 pKa = 10.51LSTPVTLVQLEE443 pKa = 4.56HH444 pKa = 6.44PTLLRR449 pKa = 11.84EE450 pKa = 4.11RR451 pKa = 11.84FTSYY455 pKa = 10.73RR456 pKa = 11.84DD457 pKa = 3.36LMGSYY462 pKa = 10.25FDD464 pKa = 4.18EE465 pKa = 4.15NVIIAVEE472 pKa = 4.08RR473 pKa = 11.84FVSLKK478 pKa = 10.38YY479 pKa = 8.06NTPMGSGYY487 pKa = 11.01AAFCYY492 pKa = 9.66IHH494 pKa = 7.1CLLSSFKK501 pKa = 11.01NFLTIFGDD509 pKa = 3.65DD510 pKa = 3.41TSIQLL515 pKa = 3.8
Molecular weight: 59.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O40962|O40962_9CLOS Putative viral coat protein OS=Little cherry virus 1 OX=217686 PE=4 SV=1
MM1 pKa = 7.19VVLVWVCLISVVCFFVSPSNIGLEE25 pKa = 4.02KK26 pKa = 10.25FSRR29 pKa = 11.84FGFSNVCC36 pKa = 3.3
MM1 pKa = 7.19VVLVWVCLISVVCFFVSPSNIGLEE25 pKa = 4.02KK26 pKa = 10.25FSRR29 pKa = 11.84FGFSNVCC36 pKa = 3.3
Molecular weight: 4.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5475 |
36 |
2301 |
608.3 |
69.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.731 ± 0.762 | 1.918 ± 0.189 |
6.557 ± 0.34 | 5.352 ± 0.54 |
6.393 ± 0.263 | 4.493 ± 0.344 |
2.009 ± 0.255 | 7.306 ± 0.403 |
7.635 ± 0.413 | 9.991 ± 0.552 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.1 ± 0.295 | 7.123 ± 0.514 |
3.087 ± 0.286 | 2.301 ± 0.219 |
4.329 ± 0.239 | 8.402 ± 0.362 |
5.205 ± 0.369 | 6.74 ± 0.467 |
0.53 ± 0.115 | 3.799 ± 0.298 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
