
Erethizon dorsatum papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
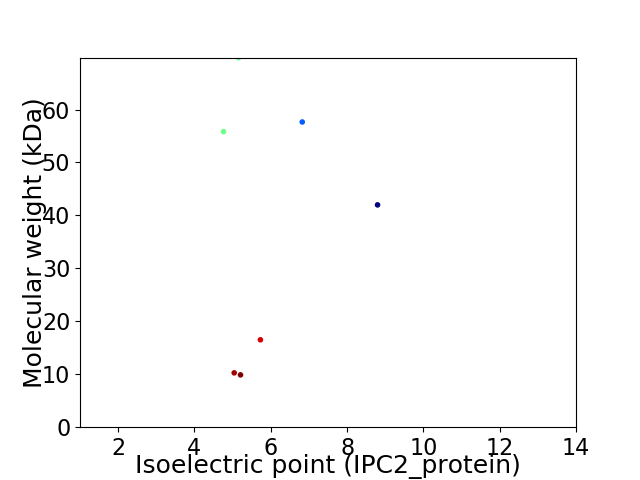
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5ENG9|A0A2Z5ENG9_9PAPI Major capsid protein L1 OS=Erethizon dorsatum papillomavirus 2 OX=2268125 GN=L1 PE=3 SV=1
MM1 pKa = 6.95VAVKK5 pKa = 10.35RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84LKK10 pKa = 10.33RR11 pKa = 11.84DD12 pKa = 3.04SPEE15 pKa = 3.92NIYY18 pKa = 10.15RR19 pKa = 11.84QCQISGNCPPDD30 pKa = 3.38VKK32 pKa = 11.17NKK34 pKa = 10.0IEE36 pKa = 4.0QNTLADD42 pKa = 5.95RR43 pKa = 11.84ILKK46 pKa = 8.59WLSSIIYY53 pKa = 10.17LGGLGIGTGRR63 pKa = 11.84GTGGSRR69 pKa = 11.84GYY71 pKa = 11.32GPINPGGRR79 pKa = 11.84VTGTGEE85 pKa = 4.22VVHH88 pKa = 6.28PAIPVDD94 pKa = 4.24PIGPTDD100 pKa = 5.11IIPVDD105 pKa = 4.1PNSSSIVPLLEE116 pKa = 4.37GGPTGDD122 pKa = 4.57VPDD125 pKa = 4.7ILPGPSGGEE134 pKa = 3.97VTSGLDD140 pKa = 3.38FASVDD145 pKa = 3.35VTTSVEE151 pKa = 4.39TIAEE155 pKa = 4.07IAPTVDD161 pKa = 2.98VTVQGAGEE169 pKa = 4.12DD170 pKa = 3.4TGATVVRR177 pKa = 11.84PSTSRR182 pKa = 11.84GATSTFFNPAYY193 pKa = 9.99VSSIQPPAIAGEE205 pKa = 4.23VGGEE209 pKa = 3.77ISGLDD214 pKa = 3.46VTDD217 pKa = 4.53LSGFTAATFDD227 pKa = 3.56RR228 pKa = 11.84AGLQEE233 pKa = 4.89EE234 pKa = 4.75IEE236 pKa = 4.35LMDD239 pKa = 3.69IVDD242 pKa = 4.44LEE244 pKa = 4.6GEE246 pKa = 4.45GVFDD250 pKa = 3.75IFEE253 pKa = 4.67PPSTSTPRR261 pKa = 11.84NSLQRR266 pKa = 11.84LIGRR270 pKa = 11.84ARR272 pKa = 11.84QLYY275 pKa = 8.74NRR277 pKa = 11.84RR278 pKa = 11.84IRR280 pKa = 11.84QQPVRR285 pKa = 11.84NPLFIQRR292 pKa = 11.84PQQLVTFEE300 pKa = 4.38YY301 pKa = 10.32EE302 pKa = 3.88NPAFSDD308 pKa = 3.97PEE310 pKa = 3.75VTMIFEE316 pKa = 4.36RR317 pKa = 11.84DD318 pKa = 3.46VAEE321 pKa = 4.21VQIPPDD327 pKa = 3.87FDD329 pKa = 4.1FLDD332 pKa = 3.82VARR335 pKa = 11.84LGRR338 pKa = 11.84PYY340 pKa = 10.81LSEE343 pKa = 3.77TADD346 pKa = 3.37RR347 pKa = 11.84TVRR350 pKa = 11.84VSRR353 pKa = 11.84LGQRR357 pKa = 11.84SSLQTRR363 pKa = 11.84QGTVVGEE370 pKa = 4.14RR371 pKa = 11.84VHH373 pKa = 6.88YY374 pKa = 10.44YY375 pKa = 10.91YY376 pKa = 10.96DD377 pKa = 4.32LSPIAAVEE385 pKa = 4.17SSSEE389 pKa = 4.23SIEE392 pKa = 4.8LNTLSAAPIQEE403 pKa = 4.22GTVDD407 pKa = 3.38TSIDD411 pKa = 3.55NSSALLLDD419 pKa = 3.63VFEE422 pKa = 5.45EE423 pKa = 4.39DD424 pKa = 3.85FSNSRR429 pKa = 11.84LHH431 pKa = 6.28IPLYY435 pKa = 10.85AGTRR439 pKa = 11.84NYY441 pKa = 10.86NVLEE445 pKa = 4.17IPLVNRR451 pKa = 11.84PSLPLLDD458 pKa = 5.84DD459 pKa = 3.63SGIWHH464 pKa = 7.49LSGAPAGMAVQSSSEE479 pKa = 3.89GDD481 pKa = 3.38TTLVTVPFTSVDD493 pKa = 3.91YY494 pKa = 10.27YY495 pKa = 11.04LHH497 pKa = 6.84PSLVKK502 pKa = 9.46RR503 pKa = 11.84RR504 pKa = 11.84RR505 pKa = 11.84RR506 pKa = 11.84RR507 pKa = 11.84RR508 pKa = 11.84RR509 pKa = 11.84PFGDD513 pKa = 4.01FFF515 pKa = 6.52
MM1 pKa = 6.95VAVKK5 pKa = 10.35RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84LKK10 pKa = 10.33RR11 pKa = 11.84DD12 pKa = 3.04SPEE15 pKa = 3.92NIYY18 pKa = 10.15RR19 pKa = 11.84QCQISGNCPPDD30 pKa = 3.38VKK32 pKa = 11.17NKK34 pKa = 10.0IEE36 pKa = 4.0QNTLADD42 pKa = 5.95RR43 pKa = 11.84ILKK46 pKa = 8.59WLSSIIYY53 pKa = 10.17LGGLGIGTGRR63 pKa = 11.84GTGGSRR69 pKa = 11.84GYY71 pKa = 11.32GPINPGGRR79 pKa = 11.84VTGTGEE85 pKa = 4.22VVHH88 pKa = 6.28PAIPVDD94 pKa = 4.24PIGPTDD100 pKa = 5.11IIPVDD105 pKa = 4.1PNSSSIVPLLEE116 pKa = 4.37GGPTGDD122 pKa = 4.57VPDD125 pKa = 4.7ILPGPSGGEE134 pKa = 3.97VTSGLDD140 pKa = 3.38FASVDD145 pKa = 3.35VTTSVEE151 pKa = 4.39TIAEE155 pKa = 4.07IAPTVDD161 pKa = 2.98VTVQGAGEE169 pKa = 4.12DD170 pKa = 3.4TGATVVRR177 pKa = 11.84PSTSRR182 pKa = 11.84GATSTFFNPAYY193 pKa = 9.99VSSIQPPAIAGEE205 pKa = 4.23VGGEE209 pKa = 3.77ISGLDD214 pKa = 3.46VTDD217 pKa = 4.53LSGFTAATFDD227 pKa = 3.56RR228 pKa = 11.84AGLQEE233 pKa = 4.89EE234 pKa = 4.75IEE236 pKa = 4.35LMDD239 pKa = 3.69IVDD242 pKa = 4.44LEE244 pKa = 4.6GEE246 pKa = 4.45GVFDD250 pKa = 3.75IFEE253 pKa = 4.67PPSTSTPRR261 pKa = 11.84NSLQRR266 pKa = 11.84LIGRR270 pKa = 11.84ARR272 pKa = 11.84QLYY275 pKa = 8.74NRR277 pKa = 11.84RR278 pKa = 11.84IRR280 pKa = 11.84QQPVRR285 pKa = 11.84NPLFIQRR292 pKa = 11.84PQQLVTFEE300 pKa = 4.38YY301 pKa = 10.32EE302 pKa = 3.88NPAFSDD308 pKa = 3.97PEE310 pKa = 3.75VTMIFEE316 pKa = 4.36RR317 pKa = 11.84DD318 pKa = 3.46VAEE321 pKa = 4.21VQIPPDD327 pKa = 3.87FDD329 pKa = 4.1FLDD332 pKa = 3.82VARR335 pKa = 11.84LGRR338 pKa = 11.84PYY340 pKa = 10.81LSEE343 pKa = 3.77TADD346 pKa = 3.37RR347 pKa = 11.84TVRR350 pKa = 11.84VSRR353 pKa = 11.84LGQRR357 pKa = 11.84SSLQTRR363 pKa = 11.84QGTVVGEE370 pKa = 4.14RR371 pKa = 11.84VHH373 pKa = 6.88YY374 pKa = 10.44YY375 pKa = 10.91YY376 pKa = 10.96DD377 pKa = 4.32LSPIAAVEE385 pKa = 4.17SSSEE389 pKa = 4.23SIEE392 pKa = 4.8LNTLSAAPIQEE403 pKa = 4.22GTVDD407 pKa = 3.38TSIDD411 pKa = 3.55NSSALLLDD419 pKa = 3.63VFEE422 pKa = 5.45EE423 pKa = 4.39DD424 pKa = 3.85FSNSRR429 pKa = 11.84LHH431 pKa = 6.28IPLYY435 pKa = 10.85AGTRR439 pKa = 11.84NYY441 pKa = 10.86NVLEE445 pKa = 4.17IPLVNRR451 pKa = 11.84PSLPLLDD458 pKa = 5.84DD459 pKa = 3.63SGIWHH464 pKa = 7.49LSGAPAGMAVQSSSEE479 pKa = 3.89GDD481 pKa = 3.38TTLVTVPFTSVDD493 pKa = 3.91YY494 pKa = 10.27YY495 pKa = 11.04LHH497 pKa = 6.84PSLVKK502 pKa = 9.46RR503 pKa = 11.84RR504 pKa = 11.84RR505 pKa = 11.84RR506 pKa = 11.84RR507 pKa = 11.84RR508 pKa = 11.84RR509 pKa = 11.84PFGDD513 pKa = 4.01FFF515 pKa = 6.52
Molecular weight: 55.82 kDa
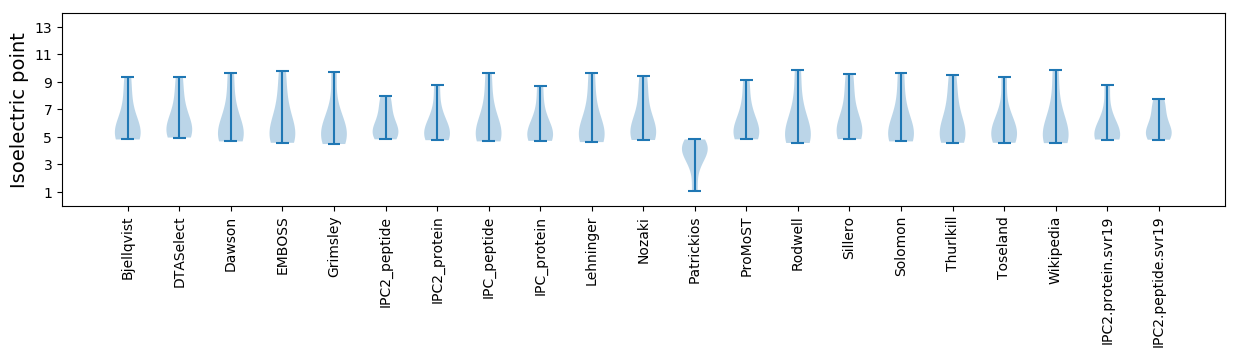
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5END3|A0A2Z5END3_9PAPI Protein E6 OS=Erethizon dorsatum papillomavirus 2 OX=2268125 GN=E6 PE=3 SV=1
MM1 pKa = 6.4QTLEE5 pKa = 3.78GRR7 pKa = 11.84YY8 pKa = 9.71NLLQEE13 pKa = 3.96QLLNLFEE20 pKa = 5.24KK21 pKa = 10.82GSSEE25 pKa = 5.41LEE27 pKa = 3.97DD28 pKa = 3.93QIQYY32 pKa = 10.22WDD34 pKa = 3.88LQRR37 pKa = 11.84QEE39 pKa = 4.78QILLHH44 pKa = 5.31YY45 pKa = 10.03ARR47 pKa = 11.84KK48 pKa = 9.51QGRR51 pKa = 11.84SHH53 pKa = 7.91IGMVTVPALSTSEE66 pKa = 4.14YY67 pKa = 8.47NAKK70 pKa = 10.18VAIKK74 pKa = 10.58VGLLLKK80 pKa = 10.6SLAKK84 pKa = 9.87SRR86 pKa = 11.84YY87 pKa = 8.37GRR89 pKa = 11.84EE90 pKa = 3.79PWSMQEE96 pKa = 3.95TSSEE100 pKa = 4.13LVLDD104 pKa = 4.47TKK106 pKa = 10.99PKK108 pKa = 10.96GLFKK112 pKa = 10.99KK113 pKa = 10.25EE114 pKa = 3.88GTHH117 pKa = 6.01VDD119 pKa = 2.59VWYY122 pKa = 10.18DD123 pKa = 3.4KK124 pKa = 11.25DD125 pKa = 3.86PEE127 pKa = 4.56NGAQYY132 pKa = 10.87VLWKK136 pKa = 10.39YY137 pKa = 10.47LYY139 pKa = 10.69KK140 pKa = 10.08DD141 pKa = 3.55TEE143 pKa = 4.43YY144 pKa = 11.39GWVKK148 pKa = 10.54LKK150 pKa = 10.92SHH152 pKa = 6.25VDD154 pKa = 3.17YY155 pKa = 11.33HH156 pKa = 6.43GIYY159 pKa = 10.64YY160 pKa = 10.17KK161 pKa = 10.91DD162 pKa = 3.39EE163 pKa = 4.25TDD165 pKa = 2.99EE166 pKa = 4.63KK167 pKa = 10.6IYY169 pKa = 9.74YY170 pKa = 9.82TSFDD174 pKa = 3.81EE175 pKa = 5.46DD176 pKa = 3.59GVRR179 pKa = 11.84YY180 pKa = 8.22STTGTWIVHH189 pKa = 5.87FQNDD193 pKa = 4.46TISSSRR199 pKa = 11.84EE200 pKa = 3.37EE201 pKa = 4.06RR202 pKa = 11.84DD203 pKa = 3.19RR204 pKa = 11.84SEE206 pKa = 4.28TTGSAGTEE214 pKa = 4.06TSRR217 pKa = 11.84SEE219 pKa = 4.19RR220 pKa = 11.84PPPPRR225 pKa = 11.84SRR227 pKa = 11.84GSAGGGGGEE236 pKa = 3.74RR237 pKa = 11.84RR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84SIEE243 pKa = 3.57QAPNPTEE250 pKa = 3.7VGQRR254 pKa = 11.84RR255 pKa = 11.84TSVPKK260 pKa = 10.63HH261 pKa = 4.85NLSRR265 pKa = 11.84LKK267 pKa = 10.55RR268 pKa = 11.84LNLEE272 pKa = 4.04ARR274 pKa = 11.84DD275 pKa = 4.0PPVLILKK282 pKa = 8.94GQANALKK289 pKa = 10.06CWRR292 pKa = 11.84HH293 pKa = 4.12RR294 pKa = 11.84QKK296 pKa = 10.83CKK298 pKa = 10.08RR299 pKa = 11.84EE300 pKa = 3.8KK301 pKa = 10.85LFVDD305 pKa = 4.14ISTTFAWSGCGVGVGAGAGARR326 pKa = 11.84KK327 pKa = 9.2NRR329 pKa = 11.84QINSRR334 pKa = 11.84LLVAFADD341 pKa = 3.87STHH344 pKa = 7.08RR345 pKa = 11.84DD346 pKa = 3.41LFLKK350 pKa = 10.28KK351 pKa = 9.98IPIPKK356 pKa = 9.85GCTYY360 pKa = 10.92DD361 pKa = 3.56YY362 pKa = 11.63GNLSSLL368 pKa = 3.91
MM1 pKa = 6.4QTLEE5 pKa = 3.78GRR7 pKa = 11.84YY8 pKa = 9.71NLLQEE13 pKa = 3.96QLLNLFEE20 pKa = 5.24KK21 pKa = 10.82GSSEE25 pKa = 5.41LEE27 pKa = 3.97DD28 pKa = 3.93QIQYY32 pKa = 10.22WDD34 pKa = 3.88LQRR37 pKa = 11.84QEE39 pKa = 4.78QILLHH44 pKa = 5.31YY45 pKa = 10.03ARR47 pKa = 11.84KK48 pKa = 9.51QGRR51 pKa = 11.84SHH53 pKa = 7.91IGMVTVPALSTSEE66 pKa = 4.14YY67 pKa = 8.47NAKK70 pKa = 10.18VAIKK74 pKa = 10.58VGLLLKK80 pKa = 10.6SLAKK84 pKa = 9.87SRR86 pKa = 11.84YY87 pKa = 8.37GRR89 pKa = 11.84EE90 pKa = 3.79PWSMQEE96 pKa = 3.95TSSEE100 pKa = 4.13LVLDD104 pKa = 4.47TKK106 pKa = 10.99PKK108 pKa = 10.96GLFKK112 pKa = 10.99KK113 pKa = 10.25EE114 pKa = 3.88GTHH117 pKa = 6.01VDD119 pKa = 2.59VWYY122 pKa = 10.18DD123 pKa = 3.4KK124 pKa = 11.25DD125 pKa = 3.86PEE127 pKa = 4.56NGAQYY132 pKa = 10.87VLWKK136 pKa = 10.39YY137 pKa = 10.47LYY139 pKa = 10.69KK140 pKa = 10.08DD141 pKa = 3.55TEE143 pKa = 4.43YY144 pKa = 11.39GWVKK148 pKa = 10.54LKK150 pKa = 10.92SHH152 pKa = 6.25VDD154 pKa = 3.17YY155 pKa = 11.33HH156 pKa = 6.43GIYY159 pKa = 10.64YY160 pKa = 10.17KK161 pKa = 10.91DD162 pKa = 3.39EE163 pKa = 4.25TDD165 pKa = 2.99EE166 pKa = 4.63KK167 pKa = 10.6IYY169 pKa = 9.74YY170 pKa = 9.82TSFDD174 pKa = 3.81EE175 pKa = 5.46DD176 pKa = 3.59GVRR179 pKa = 11.84YY180 pKa = 8.22STTGTWIVHH189 pKa = 5.87FQNDD193 pKa = 4.46TISSSRR199 pKa = 11.84EE200 pKa = 3.37EE201 pKa = 4.06RR202 pKa = 11.84DD203 pKa = 3.19RR204 pKa = 11.84SEE206 pKa = 4.28TTGSAGTEE214 pKa = 4.06TSRR217 pKa = 11.84SEE219 pKa = 4.19RR220 pKa = 11.84PPPPRR225 pKa = 11.84SRR227 pKa = 11.84GSAGGGGGEE236 pKa = 3.74RR237 pKa = 11.84RR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84SIEE243 pKa = 3.57QAPNPTEE250 pKa = 3.7VGQRR254 pKa = 11.84RR255 pKa = 11.84TSVPKK260 pKa = 10.63HH261 pKa = 4.85NLSRR265 pKa = 11.84LKK267 pKa = 10.55RR268 pKa = 11.84LNLEE272 pKa = 4.04ARR274 pKa = 11.84DD275 pKa = 4.0PPVLILKK282 pKa = 8.94GQANALKK289 pKa = 10.06CWRR292 pKa = 11.84HH293 pKa = 4.12RR294 pKa = 11.84QKK296 pKa = 10.83CKK298 pKa = 10.08RR299 pKa = 11.84EE300 pKa = 3.8KK301 pKa = 10.85LFVDD305 pKa = 4.14ISTTFAWSGCGVGVGAGAGARR326 pKa = 11.84KK327 pKa = 9.2NRR329 pKa = 11.84QINSRR334 pKa = 11.84LLVAFADD341 pKa = 3.87STHH344 pKa = 7.08RR345 pKa = 11.84DD346 pKa = 3.41LFLKK350 pKa = 10.28KK351 pKa = 9.98IPIPKK356 pKa = 9.85GCTYY360 pKa = 10.92DD361 pKa = 3.56YY362 pKa = 11.63GNLSSLL368 pKa = 3.91
Molecular weight: 41.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2323 |
86 |
608 |
331.9 |
37.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.209 ± 0.48 | 2.109 ± 0.788 |
6.156 ± 0.351 | 6.457 ± 0.406 |
4.09 ± 0.589 | 6.974 ± 0.804 |
2.238 ± 0.386 | 4.649 ± 0.695 |
4.95 ± 1.025 | 9.643 ± 0.883 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.55 ± 0.403 | 4.133 ± 0.477 |
5.639 ± 1.008 | 5.08 ± 0.544 |
6.07 ± 0.77 | 7.232 ± 0.583 |
6.07 ± 0.521 | 6.802 ± 0.556 |
1.464 ± 0.31 | 3.487 ± 0.335 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
