
Beihai sobemo-like virus 14
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.4
Get precalculated fractions of proteins
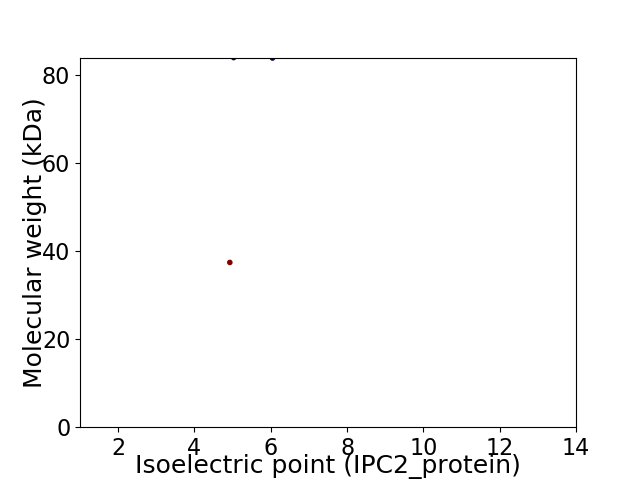
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
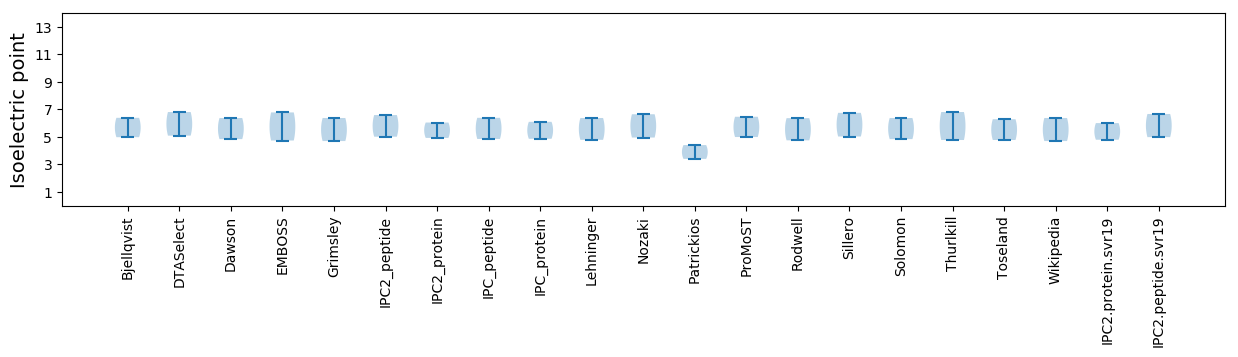
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEE5|A0A1L3KEE5_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 14 OX=1922685 PE=4 SV=1
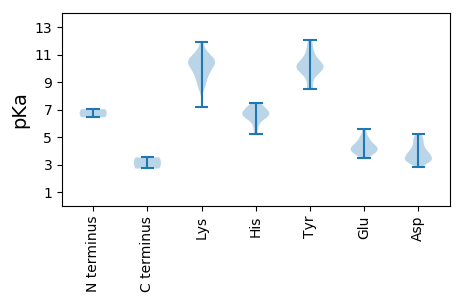
MM1 pKa = 7.03VGYY4 pKa = 10.24RR5 pKa = 11.84GNRR8 pKa = 11.84VEE10 pKa = 4.47DD11 pKa = 3.42LRR13 pKa = 11.84LAVLLRR19 pKa = 11.84LEE21 pKa = 4.29EE22 pKa = 4.12LQEE25 pKa = 4.24GKK27 pKa = 10.5ALNDD31 pKa = 3.0VFSFIKK37 pKa = 10.45QEE39 pKa = 3.77PHH41 pKa = 6.5KK42 pKa = 10.28LAKK45 pKa = 10.28AEE47 pKa = 3.81QSMWRR52 pKa = 11.84LISGLSCTDD61 pKa = 3.16QLVATLLSEE70 pKa = 4.33EE71 pKa = 3.91LMAYY75 pKa = 9.88VVDD78 pKa = 4.86HH79 pKa = 6.64PMITNMGIGWGFAMEE94 pKa = 5.34GGVSWLRR101 pKa = 11.84NFMPSNPVAADD112 pKa = 3.29KK113 pKa = 11.31SSWDD117 pKa = 3.23WTVQPWVYY125 pKa = 11.33DD126 pKa = 3.58CFLEE130 pKa = 5.0IMLWLHH136 pKa = 7.47DD137 pKa = 4.85DD138 pKa = 4.92FSTCKK143 pKa = 10.33QKK145 pKa = 10.46ILRR148 pKa = 11.84NHH150 pKa = 6.28ILSVCEE156 pKa = 3.88EE157 pKa = 4.27KK158 pKa = 11.23VFDD161 pKa = 3.86TGGGRR166 pKa = 11.84LKK168 pKa = 10.59QPAPGIVASGWKK180 pKa = 7.2WTILFNCFGQLFLHH194 pKa = 6.62YY195 pKa = 10.12YY196 pKa = 10.21ADD198 pKa = 3.84PEE200 pKa = 4.6GRR202 pKa = 11.84FPPPLTMGDD211 pKa = 3.47DD212 pKa = 4.71TIQEE216 pKa = 4.3EE217 pKa = 4.38TDD219 pKa = 3.26DD220 pKa = 5.23EE221 pKa = 4.8YY222 pKa = 10.86WCQIKK227 pKa = 8.43RR228 pKa = 11.84TGAILKK234 pKa = 8.42EE235 pKa = 4.32CQKK238 pKa = 10.97EE239 pKa = 3.9PIYY242 pKa = 10.99CSMRR246 pKa = 11.84LNDD249 pKa = 3.57WGFVPEE255 pKa = 5.08NMGKK259 pKa = 9.18YY260 pKa = 10.08CYY262 pKa = 10.2AFHH265 pKa = 7.28HH266 pKa = 6.9LDD268 pKa = 3.35EE269 pKa = 4.66EE270 pKa = 4.72LAVEE274 pKa = 4.52TLASYY279 pKa = 10.27QWLFAYY285 pKa = 9.78MPEE288 pKa = 3.74QCAAIQRR295 pKa = 11.84WLRR298 pKa = 11.84LLGGAHH304 pKa = 6.68FVTDD308 pKa = 3.69MLYY311 pKa = 8.71MQLYY315 pKa = 8.57MEE317 pKa = 4.86GRR319 pKa = 11.84TNEE322 pKa = 3.77WW323 pKa = 2.71
MM1 pKa = 7.03VGYY4 pKa = 10.24RR5 pKa = 11.84GNRR8 pKa = 11.84VEE10 pKa = 4.47DD11 pKa = 3.42LRR13 pKa = 11.84LAVLLRR19 pKa = 11.84LEE21 pKa = 4.29EE22 pKa = 4.12LQEE25 pKa = 4.24GKK27 pKa = 10.5ALNDD31 pKa = 3.0VFSFIKK37 pKa = 10.45QEE39 pKa = 3.77PHH41 pKa = 6.5KK42 pKa = 10.28LAKK45 pKa = 10.28AEE47 pKa = 3.81QSMWRR52 pKa = 11.84LISGLSCTDD61 pKa = 3.16QLVATLLSEE70 pKa = 4.33EE71 pKa = 3.91LMAYY75 pKa = 9.88VVDD78 pKa = 4.86HH79 pKa = 6.64PMITNMGIGWGFAMEE94 pKa = 5.34GGVSWLRR101 pKa = 11.84NFMPSNPVAADD112 pKa = 3.29KK113 pKa = 11.31SSWDD117 pKa = 3.23WTVQPWVYY125 pKa = 11.33DD126 pKa = 3.58CFLEE130 pKa = 5.0IMLWLHH136 pKa = 7.47DD137 pKa = 4.85DD138 pKa = 4.92FSTCKK143 pKa = 10.33QKK145 pKa = 10.46ILRR148 pKa = 11.84NHH150 pKa = 6.28ILSVCEE156 pKa = 3.88EE157 pKa = 4.27KK158 pKa = 11.23VFDD161 pKa = 3.86TGGGRR166 pKa = 11.84LKK168 pKa = 10.59QPAPGIVASGWKK180 pKa = 7.2WTILFNCFGQLFLHH194 pKa = 6.62YY195 pKa = 10.12YY196 pKa = 10.21ADD198 pKa = 3.84PEE200 pKa = 4.6GRR202 pKa = 11.84FPPPLTMGDD211 pKa = 3.47DD212 pKa = 4.71TIQEE216 pKa = 4.3EE217 pKa = 4.38TDD219 pKa = 3.26DD220 pKa = 5.23EE221 pKa = 4.8YY222 pKa = 10.86WCQIKK227 pKa = 8.43RR228 pKa = 11.84TGAILKK234 pKa = 8.42EE235 pKa = 4.32CQKK238 pKa = 10.97EE239 pKa = 3.9PIYY242 pKa = 10.99CSMRR246 pKa = 11.84LNDD249 pKa = 3.57WGFVPEE255 pKa = 5.08NMGKK259 pKa = 9.18YY260 pKa = 10.08CYY262 pKa = 10.2AFHH265 pKa = 7.28HH266 pKa = 6.9LDD268 pKa = 3.35EE269 pKa = 4.66EE270 pKa = 4.72LAVEE274 pKa = 4.52TLASYY279 pKa = 10.27QWLFAYY285 pKa = 9.78MPEE288 pKa = 3.74QCAAIQRR295 pKa = 11.84WLRR298 pKa = 11.84LLGGAHH304 pKa = 6.68FVTDD308 pKa = 3.69MLYY311 pKa = 8.71MQLYY315 pKa = 8.57MEE317 pKa = 4.86GRR319 pKa = 11.84TNEE322 pKa = 3.77WW323 pKa = 2.71
Molecular weight: 37.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEE5|A0A1L3KEE5_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 14 OX=1922685 PE=4 SV=1
MM1 pKa = 6.44WWARR5 pKa = 11.84SLYY8 pKa = 9.34LTIFGLIVQVSRR20 pKa = 11.84SGPSQSLVSFLEE32 pKa = 4.13TGSYY36 pKa = 10.15KK37 pKa = 10.81SFWKK41 pKa = 10.55VPGYY45 pKa = 10.1QEE47 pKa = 4.76VEE49 pKa = 4.01YY50 pKa = 10.1STDD53 pKa = 3.27NNTKK57 pKa = 9.19SVCSVGLMPEE67 pKa = 4.63GPRR70 pKa = 11.84GSFYY74 pKa = 10.98HH75 pKa = 6.78LGCATCVLTQVKK87 pKa = 9.98RR88 pKa = 11.84DD89 pKa = 3.44GVLADD94 pKa = 3.9EE95 pKa = 5.2FLTATHH101 pKa = 5.22VTEE104 pKa = 4.48DD105 pKa = 3.78QKK107 pKa = 11.86DD108 pKa = 3.76LYY110 pKa = 9.88VWRR113 pKa = 11.84NGRR116 pKa = 11.84VAKK119 pKa = 10.35VKK121 pKa = 9.93QFWQPAGQKK130 pKa = 9.8CFYY133 pKa = 9.9EE134 pKa = 4.27IMILRR139 pKa = 11.84VVSGASLLGCKK150 pKa = 8.72PAKK153 pKa = 9.36TVSSFKK159 pKa = 10.69DD160 pKa = 3.42DD161 pKa = 4.58NIVNLYY167 pKa = 10.29AAVEE171 pKa = 4.05GRR173 pKa = 11.84LIKK176 pKa = 8.65TTGTIASMGKK186 pKa = 9.52SAMLGIAHH194 pKa = 7.13TANTLPGWSGAGIFTPSGLVAFHH217 pKa = 6.03HH218 pKa = 6.54HH219 pKa = 6.9GIKK222 pKa = 10.23KK223 pKa = 8.72EE224 pKa = 3.96AKK226 pKa = 9.75DD227 pKa = 3.59GKK229 pKa = 10.38GCINTGYY236 pKa = 10.45EE237 pKa = 3.48IGSIINLLRR246 pKa = 11.84RR247 pKa = 11.84SAEE250 pKa = 3.75ARR252 pKa = 11.84PKK254 pKa = 10.38LNQLVTHH261 pKa = 7.12DD262 pKa = 4.14NQLSSEE268 pKa = 4.34CEE270 pKa = 4.84SVDD273 pKa = 2.83TWNWFKK279 pKa = 11.9DD280 pKa = 3.31MVEE283 pKa = 3.92AGKK286 pKa = 9.12MEE288 pKa = 4.19MTVTPWGDD296 pKa = 3.2EE297 pKa = 3.65QLYY300 pKa = 8.51TFTHH304 pKa = 5.66SGKK307 pKa = 9.58RR308 pKa = 11.84WMYY311 pKa = 10.53TSDD314 pKa = 3.85EE315 pKa = 4.49LPDD318 pKa = 4.55DD319 pKa = 4.23FLDD322 pKa = 4.96EE323 pKa = 4.3IEE325 pKa = 5.04EE326 pKa = 4.25YY327 pKa = 11.21ADD329 pKa = 4.42DD330 pKa = 4.26SLIDD334 pKa = 3.88EE335 pKa = 5.04EE336 pKa = 5.5DD337 pKa = 4.46DD338 pKa = 3.31YY339 pKa = 11.7HH340 pKa = 7.18QRR342 pKa = 11.84SRR344 pKa = 11.84GKK346 pKa = 8.83RR347 pKa = 11.84TSSRR351 pKa = 11.84YY352 pKa = 8.65RR353 pKa = 11.84DD354 pKa = 3.49EE355 pKa = 5.56LGRR358 pKa = 11.84CSDD361 pKa = 3.68DD362 pKa = 3.84DD363 pKa = 5.02GYY365 pKa = 11.98GEE367 pKa = 5.62APDD370 pKa = 4.89DD371 pKa = 5.09DD372 pKa = 4.41YY373 pKa = 12.04PEE375 pKa = 4.88IYY377 pKa = 10.49SDD379 pKa = 4.2FEE381 pKa = 4.5CVSSKK386 pKa = 11.06EE387 pKa = 4.01KK388 pKa = 10.62LDD390 pKa = 3.88PLGMPMTPIDD400 pKa = 4.11LEE402 pKa = 4.04AVEE405 pKa = 4.86ALRR408 pKa = 11.84EE409 pKa = 4.41AIDD412 pKa = 3.57EE413 pKa = 4.18VDD415 pKa = 3.12IQKK418 pKa = 11.17AKK420 pKa = 9.03VTAFLRR426 pKa = 11.84ALSEE430 pKa = 4.06AQIKK434 pKa = 9.54ARR436 pKa = 11.84RR437 pKa = 11.84AWEE440 pKa = 3.99ADD442 pKa = 3.2EE443 pKa = 4.54TDD445 pKa = 3.23AKK447 pKa = 10.89FDD449 pKa = 4.17VILQASADD457 pKa = 3.88IDD459 pKa = 3.92CLQAFCEE466 pKa = 4.06RR467 pKa = 11.84FAAEE471 pKa = 4.31RR472 pKa = 11.84EE473 pKa = 4.45SFTAKK478 pKa = 10.41YY479 pKa = 10.06NEE481 pKa = 3.95VMQKK485 pKa = 10.65SDD487 pKa = 3.24VDD489 pKa = 3.77STLDD493 pKa = 3.19RR494 pKa = 11.84RR495 pKa = 11.84VARR498 pKa = 11.84KK499 pKa = 9.86KK500 pKa = 10.28KK501 pKa = 10.28GKK503 pKa = 9.9EE504 pKa = 3.64KK505 pKa = 10.68LPPLPYY511 pKa = 10.06TSGLRR516 pKa = 11.84DD517 pKa = 3.53DD518 pKa = 4.46AVRR521 pKa = 11.84SIDD524 pKa = 3.29VCRR527 pKa = 11.84AVVDD531 pKa = 4.84SIPMRR536 pKa = 11.84NRR538 pKa = 11.84ASAAGVEE545 pKa = 4.59VQVSQPEE552 pKa = 4.1GLYY555 pKa = 9.45TDD557 pKa = 3.67EE558 pKa = 4.14GRR560 pKa = 11.84IVLAEE565 pKa = 4.03VYY567 pKa = 9.65GVKK570 pKa = 10.28VKK572 pKa = 11.01SDD574 pKa = 3.01IKK576 pKa = 10.79RR577 pKa = 11.84DD578 pKa = 3.2IKK580 pKa = 11.35GPLNYY585 pKa = 9.88KK586 pKa = 9.62GAPQRR591 pKa = 11.84LEE593 pKa = 3.78MSPDD597 pKa = 3.33GDD599 pKa = 3.91SQRR602 pKa = 11.84DD603 pKa = 3.42SSRR606 pKa = 11.84ASEE609 pKa = 4.2KK610 pKa = 9.7VQKK613 pKa = 10.64SSLITSTIQSKK624 pKa = 10.3EE625 pKa = 3.97NQEE628 pKa = 3.88NTQMSRR634 pKa = 11.84ISSGNCSQQQTNLLPPRR651 pKa = 11.84RR652 pKa = 11.84RR653 pKa = 11.84GTGEE657 pKa = 3.61RR658 pKa = 11.84LKK660 pKa = 10.8SHH662 pKa = 6.39SEE664 pKa = 4.3SILSEE669 pKa = 3.75HH670 pKa = 6.59GKK672 pKa = 9.32PVIAASHH679 pKa = 6.74LIEE682 pKa = 5.45LYY684 pKa = 10.42RR685 pKa = 11.84EE686 pKa = 4.09PLSKK690 pKa = 10.5EE691 pKa = 3.62FQSYY695 pKa = 9.8LRR697 pKa = 11.84EE698 pKa = 4.03PRR700 pKa = 11.84TVSSEE705 pKa = 3.62QEE707 pKa = 3.65RR708 pKa = 11.84RR709 pKa = 11.84KK710 pKa = 10.63KK711 pKa = 10.34KK712 pKa = 9.62IKK714 pKa = 10.05NVRR717 pKa = 11.84DD718 pKa = 3.43VANPRR723 pKa = 11.84ASSKK727 pKa = 11.0NFLSSAAPSVSRR739 pKa = 11.84AGFRR743 pKa = 11.84GPVANKK749 pKa = 9.92RR750 pKa = 11.84CC751 pKa = 3.52
MM1 pKa = 6.44WWARR5 pKa = 11.84SLYY8 pKa = 9.34LTIFGLIVQVSRR20 pKa = 11.84SGPSQSLVSFLEE32 pKa = 4.13TGSYY36 pKa = 10.15KK37 pKa = 10.81SFWKK41 pKa = 10.55VPGYY45 pKa = 10.1QEE47 pKa = 4.76VEE49 pKa = 4.01YY50 pKa = 10.1STDD53 pKa = 3.27NNTKK57 pKa = 9.19SVCSVGLMPEE67 pKa = 4.63GPRR70 pKa = 11.84GSFYY74 pKa = 10.98HH75 pKa = 6.78LGCATCVLTQVKK87 pKa = 9.98RR88 pKa = 11.84DD89 pKa = 3.44GVLADD94 pKa = 3.9EE95 pKa = 5.2FLTATHH101 pKa = 5.22VTEE104 pKa = 4.48DD105 pKa = 3.78QKK107 pKa = 11.86DD108 pKa = 3.76LYY110 pKa = 9.88VWRR113 pKa = 11.84NGRR116 pKa = 11.84VAKK119 pKa = 10.35VKK121 pKa = 9.93QFWQPAGQKK130 pKa = 9.8CFYY133 pKa = 9.9EE134 pKa = 4.27IMILRR139 pKa = 11.84VVSGASLLGCKK150 pKa = 8.72PAKK153 pKa = 9.36TVSSFKK159 pKa = 10.69DD160 pKa = 3.42DD161 pKa = 4.58NIVNLYY167 pKa = 10.29AAVEE171 pKa = 4.05GRR173 pKa = 11.84LIKK176 pKa = 8.65TTGTIASMGKK186 pKa = 9.52SAMLGIAHH194 pKa = 7.13TANTLPGWSGAGIFTPSGLVAFHH217 pKa = 6.03HH218 pKa = 6.54HH219 pKa = 6.9GIKK222 pKa = 10.23KK223 pKa = 8.72EE224 pKa = 3.96AKK226 pKa = 9.75DD227 pKa = 3.59GKK229 pKa = 10.38GCINTGYY236 pKa = 10.45EE237 pKa = 3.48IGSIINLLRR246 pKa = 11.84RR247 pKa = 11.84SAEE250 pKa = 3.75ARR252 pKa = 11.84PKK254 pKa = 10.38LNQLVTHH261 pKa = 7.12DD262 pKa = 4.14NQLSSEE268 pKa = 4.34CEE270 pKa = 4.84SVDD273 pKa = 2.83TWNWFKK279 pKa = 11.9DD280 pKa = 3.31MVEE283 pKa = 3.92AGKK286 pKa = 9.12MEE288 pKa = 4.19MTVTPWGDD296 pKa = 3.2EE297 pKa = 3.65QLYY300 pKa = 8.51TFTHH304 pKa = 5.66SGKK307 pKa = 9.58RR308 pKa = 11.84WMYY311 pKa = 10.53TSDD314 pKa = 3.85EE315 pKa = 4.49LPDD318 pKa = 4.55DD319 pKa = 4.23FLDD322 pKa = 4.96EE323 pKa = 4.3IEE325 pKa = 5.04EE326 pKa = 4.25YY327 pKa = 11.21ADD329 pKa = 4.42DD330 pKa = 4.26SLIDD334 pKa = 3.88EE335 pKa = 5.04EE336 pKa = 5.5DD337 pKa = 4.46DD338 pKa = 3.31YY339 pKa = 11.7HH340 pKa = 7.18QRR342 pKa = 11.84SRR344 pKa = 11.84GKK346 pKa = 8.83RR347 pKa = 11.84TSSRR351 pKa = 11.84YY352 pKa = 8.65RR353 pKa = 11.84DD354 pKa = 3.49EE355 pKa = 5.56LGRR358 pKa = 11.84CSDD361 pKa = 3.68DD362 pKa = 3.84DD363 pKa = 5.02GYY365 pKa = 11.98GEE367 pKa = 5.62APDD370 pKa = 4.89DD371 pKa = 5.09DD372 pKa = 4.41YY373 pKa = 12.04PEE375 pKa = 4.88IYY377 pKa = 10.49SDD379 pKa = 4.2FEE381 pKa = 4.5CVSSKK386 pKa = 11.06EE387 pKa = 4.01KK388 pKa = 10.62LDD390 pKa = 3.88PLGMPMTPIDD400 pKa = 4.11LEE402 pKa = 4.04AVEE405 pKa = 4.86ALRR408 pKa = 11.84EE409 pKa = 4.41AIDD412 pKa = 3.57EE413 pKa = 4.18VDD415 pKa = 3.12IQKK418 pKa = 11.17AKK420 pKa = 9.03VTAFLRR426 pKa = 11.84ALSEE430 pKa = 4.06AQIKK434 pKa = 9.54ARR436 pKa = 11.84RR437 pKa = 11.84AWEE440 pKa = 3.99ADD442 pKa = 3.2EE443 pKa = 4.54TDD445 pKa = 3.23AKK447 pKa = 10.89FDD449 pKa = 4.17VILQASADD457 pKa = 3.88IDD459 pKa = 3.92CLQAFCEE466 pKa = 4.06RR467 pKa = 11.84FAAEE471 pKa = 4.31RR472 pKa = 11.84EE473 pKa = 4.45SFTAKK478 pKa = 10.41YY479 pKa = 10.06NEE481 pKa = 3.95VMQKK485 pKa = 10.65SDD487 pKa = 3.24VDD489 pKa = 3.77STLDD493 pKa = 3.19RR494 pKa = 11.84RR495 pKa = 11.84VARR498 pKa = 11.84KK499 pKa = 9.86KK500 pKa = 10.28KK501 pKa = 10.28GKK503 pKa = 9.9EE504 pKa = 3.64KK505 pKa = 10.68LPPLPYY511 pKa = 10.06TSGLRR516 pKa = 11.84DD517 pKa = 3.53DD518 pKa = 4.46AVRR521 pKa = 11.84SIDD524 pKa = 3.29VCRR527 pKa = 11.84AVVDD531 pKa = 4.84SIPMRR536 pKa = 11.84NRR538 pKa = 11.84ASAAGVEE545 pKa = 4.59VQVSQPEE552 pKa = 4.1GLYY555 pKa = 9.45TDD557 pKa = 3.67EE558 pKa = 4.14GRR560 pKa = 11.84IVLAEE565 pKa = 4.03VYY567 pKa = 9.65GVKK570 pKa = 10.28VKK572 pKa = 11.01SDD574 pKa = 3.01IKK576 pKa = 10.79RR577 pKa = 11.84DD578 pKa = 3.2IKK580 pKa = 11.35GPLNYY585 pKa = 9.88KK586 pKa = 9.62GAPQRR591 pKa = 11.84LEE593 pKa = 3.78MSPDD597 pKa = 3.33GDD599 pKa = 3.91SQRR602 pKa = 11.84DD603 pKa = 3.42SSRR606 pKa = 11.84ASEE609 pKa = 4.2KK610 pKa = 9.7VQKK613 pKa = 10.64SSLITSTIQSKK624 pKa = 10.3EE625 pKa = 3.97NQEE628 pKa = 3.88NTQMSRR634 pKa = 11.84ISSGNCSQQQTNLLPPRR651 pKa = 11.84RR652 pKa = 11.84RR653 pKa = 11.84GTGEE657 pKa = 3.61RR658 pKa = 11.84LKK660 pKa = 10.8SHH662 pKa = 6.39SEE664 pKa = 4.3SILSEE669 pKa = 3.75HH670 pKa = 6.59GKK672 pKa = 9.32PVIAASHH679 pKa = 6.74LIEE682 pKa = 5.45LYY684 pKa = 10.42RR685 pKa = 11.84EE686 pKa = 4.09PLSKK690 pKa = 10.5EE691 pKa = 3.62FQSYY695 pKa = 9.8LRR697 pKa = 11.84EE698 pKa = 4.03PRR700 pKa = 11.84TVSSEE705 pKa = 3.62QEE707 pKa = 3.65RR708 pKa = 11.84RR709 pKa = 11.84KK710 pKa = 10.63KK711 pKa = 10.34KK712 pKa = 9.62IKK714 pKa = 10.05NVRR717 pKa = 11.84DD718 pKa = 3.43VANPRR723 pKa = 11.84ASSKK727 pKa = 11.0NFLSSAAPSVSRR739 pKa = 11.84AGFRR743 pKa = 11.84GPVANKK749 pKa = 9.92RR750 pKa = 11.84CC751 pKa = 3.52
Molecular weight: 83.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1074 |
323 |
751 |
537.0 |
60.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.076 ± 0.436 | 2.235 ± 0.425 |
6.518 ± 0.466 | 7.542 ± 0.098 |
3.445 ± 0.591 | 6.704 ± 0.205 |
1.862 ± 0.303 | 4.562 ± 0.112 |
6.145 ± 0.893 | 8.473 ± 1.317 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.793 ± 0.912 | 2.98 ± 0.057 |
4.283 ± 0.025 | 4.19 ± 0.224 |
6.145 ± 0.893 | 8.194 ± 1.902 |
4.842 ± 0.25 | 6.238 ± 0.481 |
2.328 ± 0.989 | 3.445 ± 0.286 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
