
Shallot virus X (ShVX)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Alphaflexiviridae; Allexivirus; Acarallexivirus
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
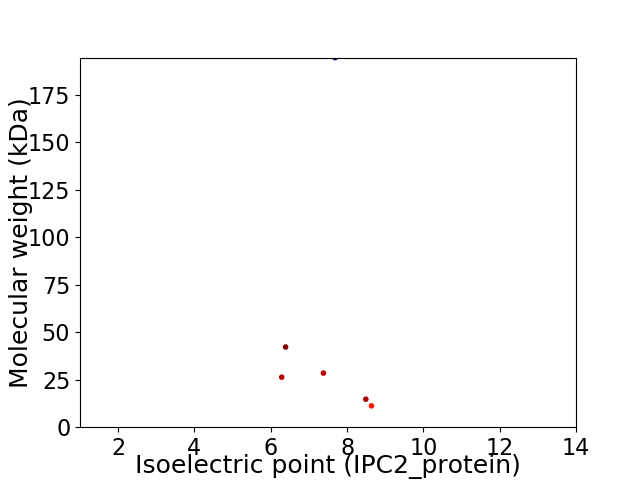
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q04582|TGB2_SHVX Movement protein TGB2 OS=Shallot virus X OX=31770 GN=ORF3 PE=3 SV=1
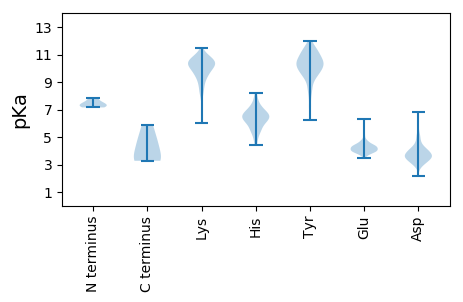
MM1 pKa = 7.15KK2 pKa = 9.91TDD4 pKa = 4.67LLLQILSNNNFTRR17 pKa = 11.84TSEE20 pKa = 4.42PIKK23 pKa = 10.5EE24 pKa = 3.92PLIIHH29 pKa = 6.47GVPGSGKK36 pKa = 8.19STLVRR41 pKa = 11.84ALVTYY46 pKa = 10.2RR47 pKa = 11.84STVACTLGAPYY58 pKa = 10.47GSNLAFPGVTSPGLTQSLTDD78 pKa = 3.41HH79 pKa = 6.21EE80 pKa = 4.5TRR82 pKa = 11.84ILDD85 pKa = 3.83EE86 pKa = 4.32YY87 pKa = 11.43QLGTEE92 pKa = 4.32SDD94 pKa = 3.68LKK96 pKa = 10.65PFNVLVGDD104 pKa = 4.27PFQGNLHH111 pKa = 6.56LKK113 pKa = 8.43AHH115 pKa = 5.15YY116 pKa = 9.97VKK118 pKa = 10.49SFSHH122 pKa = 6.12RR123 pKa = 11.84VPRR126 pKa = 11.84IICNFLQSLGYY137 pKa = 9.86EE138 pKa = 4.16IAGSKK143 pKa = 10.07PGEE146 pKa = 4.07LAQLPIYY153 pKa = 9.83GPNPSGPTGQVLHH166 pKa = 6.95LGPLSRR172 pKa = 11.84RR173 pKa = 11.84LTQSHH178 pKa = 6.1GVCSKK183 pKa = 10.85LPSEE187 pKa = 4.43VQGLEE192 pKa = 4.08FEE194 pKa = 4.58EE195 pKa = 4.56VTLVYY200 pKa = 10.42HH201 pKa = 6.47SSEE204 pKa = 4.1FEE206 pKa = 3.93RR207 pKa = 11.84NRR209 pKa = 11.84VGFYY213 pKa = 10.13IAATRR218 pKa = 11.84ALGRR222 pKa = 11.84LNLITDD228 pKa = 3.93TTLEE232 pKa = 4.46IPHH235 pKa = 6.34EE236 pKa = 4.34LCPTSS241 pKa = 3.27
MM1 pKa = 7.15KK2 pKa = 9.91TDD4 pKa = 4.67LLLQILSNNNFTRR17 pKa = 11.84TSEE20 pKa = 4.42PIKK23 pKa = 10.5EE24 pKa = 3.92PLIIHH29 pKa = 6.47GVPGSGKK36 pKa = 8.19STLVRR41 pKa = 11.84ALVTYY46 pKa = 10.2RR47 pKa = 11.84STVACTLGAPYY58 pKa = 10.47GSNLAFPGVTSPGLTQSLTDD78 pKa = 3.41HH79 pKa = 6.21EE80 pKa = 4.5TRR82 pKa = 11.84ILDD85 pKa = 3.83EE86 pKa = 4.32YY87 pKa = 11.43QLGTEE92 pKa = 4.32SDD94 pKa = 3.68LKK96 pKa = 10.65PFNVLVGDD104 pKa = 4.27PFQGNLHH111 pKa = 6.56LKK113 pKa = 8.43AHH115 pKa = 5.15YY116 pKa = 9.97VKK118 pKa = 10.49SFSHH122 pKa = 6.12RR123 pKa = 11.84VPRR126 pKa = 11.84IICNFLQSLGYY137 pKa = 9.86EE138 pKa = 4.16IAGSKK143 pKa = 10.07PGEE146 pKa = 4.07LAQLPIYY153 pKa = 9.83GPNPSGPTGQVLHH166 pKa = 6.95LGPLSRR172 pKa = 11.84RR173 pKa = 11.84LTQSHH178 pKa = 6.1GVCSKK183 pKa = 10.85LPSEE187 pKa = 4.43VQGLEE192 pKa = 4.08FEE194 pKa = 4.58EE195 pKa = 4.56VTLVYY200 pKa = 10.42HH201 pKa = 6.47SSEE204 pKa = 4.1FEE206 pKa = 3.93RR207 pKa = 11.84NRR209 pKa = 11.84VGFYY213 pKa = 10.13IAATRR218 pKa = 11.84ALGRR222 pKa = 11.84LNLITDD228 pKa = 3.93TTLEE232 pKa = 4.46IPHH235 pKa = 6.34EE236 pKa = 4.34LCPTSS241 pKa = 3.27
Molecular weight: 26.33 kDa
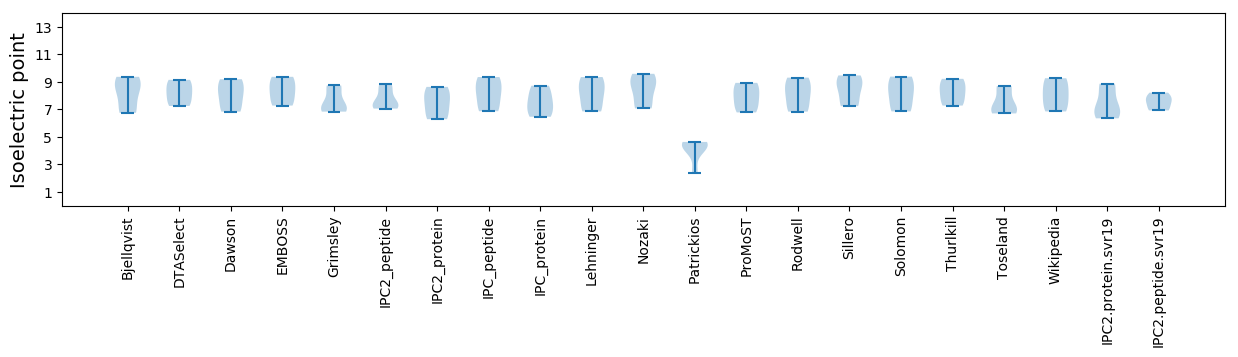
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q04583|ORF4_SHVX Uncharacterized ORF4 protein OS=Shallot virus X OX=31770 GN=ORF4 PE=4 SV=1
MM1 pKa = 7.4SFAPPPDD8 pKa = 3.29YY9 pKa = 11.38SKK11 pKa = 10.75IYY13 pKa = 9.97LALGCGLGLGFVVYY27 pKa = 10.61ASRR30 pKa = 11.84VNHH33 pKa = 6.42LPHH36 pKa = 8.21VGDD39 pKa = 3.63NTHH42 pKa = 6.49NLPHH46 pKa = 6.89GGQYY50 pKa = 10.57CDD52 pKa = 3.01GNKK55 pKa = 9.48RR56 pKa = 11.84VLYY59 pKa = 10.0SGPKK63 pKa = 9.61SGSSPTNNLWPFITVIALTLAILLTSCPRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84VCIRR98 pKa = 11.84CSQHH102 pKa = 6.04HH103 pKa = 5.88
MM1 pKa = 7.4SFAPPPDD8 pKa = 3.29YY9 pKa = 11.38SKK11 pKa = 10.75IYY13 pKa = 9.97LALGCGLGLGFVVYY27 pKa = 10.61ASRR30 pKa = 11.84VNHH33 pKa = 6.42LPHH36 pKa = 8.21VGDD39 pKa = 3.63NTHH42 pKa = 6.49NLPHH46 pKa = 6.89GGQYY50 pKa = 10.57CDD52 pKa = 3.01GNKK55 pKa = 9.48RR56 pKa = 11.84VLYY59 pKa = 10.0SGPKK63 pKa = 9.61SGSSPTNNLWPFITVIALTLAILLTSCPRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84VCIRR98 pKa = 11.84CSQHH102 pKa = 6.04HH103 pKa = 5.88
Molecular weight: 11.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2832 |
103 |
1718 |
472.0 |
52.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.309 ± 0.884 | 1.977 ± 0.616 |
5.191 ± 0.606 | 5.049 ± 0.594 |
3.919 ± 0.248 | 5.261 ± 0.825 |
3.319 ± 0.257 | 4.767 ± 0.273 |
5.544 ± 0.805 | 10.381 ± 1.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.907 ± 0.286 | 5.014 ± 0.826 |
6.321 ± 0.382 | 4.732 ± 0.451 |
5.756 ± 0.326 | 7.203 ± 0.97 |
7.556 ± 0.726 | 4.802 ± 0.438 |
0.847 ± 0.272 | 3.143 ± 0.439 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
