
Rotavirus C (isolate RVC/Human/United Kingdom/Bristol/1989) (RV-C)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus C
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
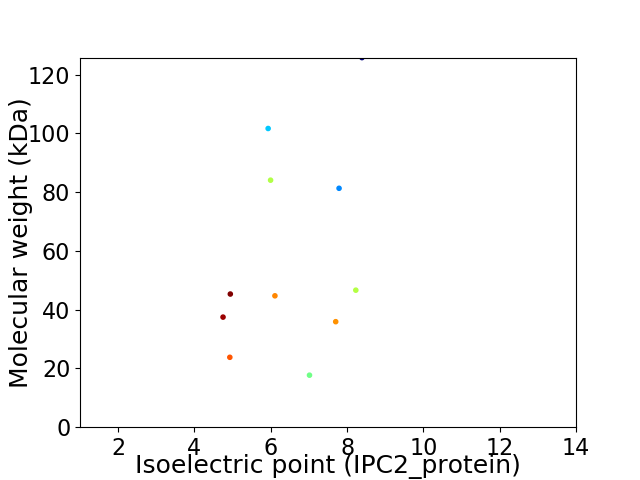
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q91E95|RDRP_ROTHC RNA-directed RNA polymerase OS=Rotavirus C (isolate RVC/Human/United Kingdom/Bristol/1989) OX=31567 PE=3 SV=1
MM1 pKa = 7.33VCTTLYY7 pKa = 8.78TVCAILFILFIYY19 pKa = 10.33ILLFRR24 pKa = 11.84KK25 pKa = 8.46MFHH28 pKa = 7.25LITDD32 pKa = 4.02TLIVILILSNCVEE45 pKa = 4.35WSQGQMFTDD54 pKa = 4.69DD55 pKa = 3.54IYY57 pKa = 11.55YY58 pKa = 10.17NGNVEE63 pKa = 4.41TIINSTDD70 pKa = 3.34PFNVEE75 pKa = 3.85SLCIYY80 pKa = 9.73FPNAVVGSQGPGKK93 pKa = 10.4SDD95 pKa = 2.77GHH97 pKa = 8.08LNDD100 pKa = 3.7GNYY103 pKa = 10.46AQTIATLFEE112 pKa = 4.43TKK114 pKa = 10.21GFPKK118 pKa = 10.58GSIILKK124 pKa = 9.46TYY126 pKa = 7.58TQTSDD131 pKa = 4.08FINSVEE137 pKa = 4.26MTCSYY142 pKa = 10.97NIVIIPDD149 pKa = 3.7SPNDD153 pKa = 3.66SEE155 pKa = 5.39SIEE158 pKa = 4.82QIAEE162 pKa = 3.94WILNVWRR169 pKa = 11.84CDD171 pKa = 3.57DD172 pKa = 4.06MNLEE176 pKa = 4.3IYY178 pKa = 9.64TYY180 pKa = 9.6EE181 pKa = 3.99QIGINNLWAAFGSDD195 pKa = 4.29CDD197 pKa = 4.41ISVCPLDD204 pKa = 3.68TTSNGIGCSPASTEE218 pKa = 4.12TYY220 pKa = 10.02EE221 pKa = 4.47VVSNDD226 pKa = 3.22TQLALINVVDD236 pKa = 4.1NVRR239 pKa = 11.84HH240 pKa = 6.38RR241 pKa = 11.84IQMNTAQCKK250 pKa = 9.81LKK252 pKa = 10.97NCIKK256 pKa = 10.93GEE258 pKa = 3.91ARR260 pKa = 11.84LNTALIRR267 pKa = 11.84ISTSSSFDD275 pKa = 3.34NSLSPLNNGQTTRR288 pKa = 11.84SFKK291 pKa = 10.82INAKK295 pKa = 9.15KK296 pKa = 9.06WWTIFYY302 pKa = 9.29TIIDD306 pKa = 4.45YY307 pKa = 11.08INTIVQAMTPRR318 pKa = 11.84HH319 pKa = 5.18RR320 pKa = 11.84AIYY323 pKa = 9.33PEE325 pKa = 3.79GWMLRR330 pKa = 11.84YY331 pKa = 9.45AA332 pKa = 4.23
MM1 pKa = 7.33VCTTLYY7 pKa = 8.78TVCAILFILFIYY19 pKa = 10.33ILLFRR24 pKa = 11.84KK25 pKa = 8.46MFHH28 pKa = 7.25LITDD32 pKa = 4.02TLIVILILSNCVEE45 pKa = 4.35WSQGQMFTDD54 pKa = 4.69DD55 pKa = 3.54IYY57 pKa = 11.55YY58 pKa = 10.17NGNVEE63 pKa = 4.41TIINSTDD70 pKa = 3.34PFNVEE75 pKa = 3.85SLCIYY80 pKa = 9.73FPNAVVGSQGPGKK93 pKa = 10.4SDD95 pKa = 2.77GHH97 pKa = 8.08LNDD100 pKa = 3.7GNYY103 pKa = 10.46AQTIATLFEE112 pKa = 4.43TKK114 pKa = 10.21GFPKK118 pKa = 10.58GSIILKK124 pKa = 9.46TYY126 pKa = 7.58TQTSDD131 pKa = 4.08FINSVEE137 pKa = 4.26MTCSYY142 pKa = 10.97NIVIIPDD149 pKa = 3.7SPNDD153 pKa = 3.66SEE155 pKa = 5.39SIEE158 pKa = 4.82QIAEE162 pKa = 3.94WILNVWRR169 pKa = 11.84CDD171 pKa = 3.57DD172 pKa = 4.06MNLEE176 pKa = 4.3IYY178 pKa = 9.64TYY180 pKa = 9.6EE181 pKa = 3.99QIGINNLWAAFGSDD195 pKa = 4.29CDD197 pKa = 4.41ISVCPLDD204 pKa = 3.68TTSNGIGCSPASTEE218 pKa = 4.12TYY220 pKa = 10.02EE221 pKa = 4.47VVSNDD226 pKa = 3.22TQLALINVVDD236 pKa = 4.1NVRR239 pKa = 11.84HH240 pKa = 6.38RR241 pKa = 11.84IQMNTAQCKK250 pKa = 9.81LKK252 pKa = 10.97NCIKK256 pKa = 10.93GEE258 pKa = 3.91ARR260 pKa = 11.84LNTALIRR267 pKa = 11.84ISTSSSFDD275 pKa = 3.34NSLSPLNNGQTTRR288 pKa = 11.84SFKK291 pKa = 10.82INAKK295 pKa = 9.15KK296 pKa = 9.06WWTIFYY302 pKa = 9.29TIIDD306 pKa = 4.45YY307 pKa = 11.08INTIVQAMTPRR318 pKa = 11.84HH319 pKa = 5.18RR320 pKa = 11.84AIYY323 pKa = 9.33PEE325 pKa = 3.79GWMLRR330 pKa = 11.84YY331 pKa = 9.45AA332 pKa = 4.23
Molecular weight: 37.47 kDa
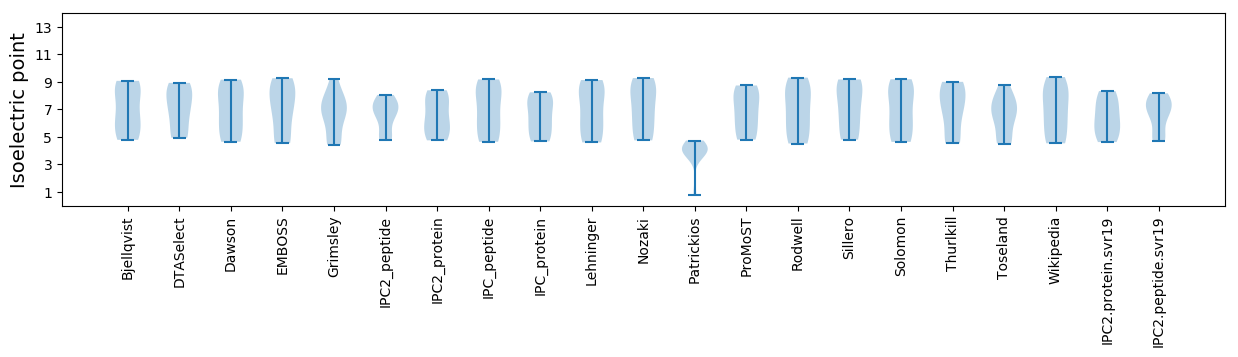
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q91E96|VP2_ROTHC Inner capsid protein VP2 OS=Rotavirus C (isolate RVC/Human/United Kingdom/Bristol/1989) OX=31567 PE=3 SV=1
MM1 pKa = 7.11AQSIVVDD8 pKa = 3.94GDD10 pKa = 3.44YY11 pKa = 11.3DD12 pKa = 3.97ALASRR17 pKa = 11.84YY18 pKa = 10.17LKK20 pKa = 10.37FVYY23 pKa = 10.25DD24 pKa = 4.32FEE26 pKa = 4.73NVTYY30 pKa = 10.35QNNYY34 pKa = 9.06FATDD38 pKa = 3.29KK39 pKa = 10.54FKK41 pKa = 11.23KK42 pKa = 10.42DD43 pKa = 2.7IEE45 pKa = 4.37QYY47 pKa = 10.67LKK49 pKa = 10.71SIHH52 pKa = 7.03DD53 pKa = 3.97GEE55 pKa = 5.93KK56 pKa = 8.92ITQSKK61 pKa = 9.92IDD63 pKa = 3.87EE64 pKa = 4.38KK65 pKa = 11.34EE66 pKa = 3.93KK67 pKa = 10.74ILLDD71 pKa = 3.42RR72 pKa = 11.84VPAEE76 pKa = 4.13EE77 pKa = 4.14RR78 pKa = 11.84CLISKK83 pKa = 9.67LVFAYY88 pKa = 9.75GKK90 pKa = 9.95HH91 pKa = 5.52GNVEE95 pKa = 4.13NKK97 pKa = 9.43LVKK100 pKa = 10.85YY101 pKa = 9.66GVKK104 pKa = 10.21DD105 pKa = 3.86ALSHH109 pKa = 7.09APQKK113 pKa = 10.15DD114 pKa = 3.2AKK116 pKa = 9.9PYY118 pKa = 8.58EE119 pKa = 4.17NNIITSEE126 pKa = 3.85IFKK129 pKa = 10.54EE130 pKa = 3.99KK131 pKa = 10.8SEE133 pKa = 4.07YY134 pKa = 9.44TDD136 pKa = 3.28IYY138 pKa = 9.73MDD140 pKa = 4.0PSINTSCQSNCQAMMFTISEE160 pKa = 4.2MKK162 pKa = 10.56LNNIKK167 pKa = 10.44NAARR171 pKa = 11.84LEE173 pKa = 4.0KK174 pKa = 10.67LFTIIAATINKK185 pKa = 9.71YY186 pKa = 10.66GMPRR190 pKa = 11.84HH191 pKa = 4.69NTRR194 pKa = 11.84YY195 pKa = 8.54RR196 pKa = 11.84YY197 pKa = 8.92EE198 pKa = 3.9WEE200 pKa = 3.94TMKK203 pKa = 10.97NKK205 pKa = 9.83PYY207 pKa = 10.77HH208 pKa = 5.31LAAWINSSIEE218 pKa = 3.9MIACVVDD225 pKa = 2.54HH226 pKa = 6.41HH227 pKa = 6.96TYY229 pKa = 9.82MIARR233 pKa = 11.84EE234 pKa = 4.43LIVKK238 pKa = 9.94SFTNRR243 pKa = 11.84TSLAKK248 pKa = 10.32LVSSPMTVLTAMLPIRR264 pKa = 11.84GTFITTEE271 pKa = 3.96NLEE274 pKa = 4.19LEE276 pKa = 4.45YY277 pKa = 11.13SNKK280 pKa = 10.12SINYY284 pKa = 8.98LISKK288 pKa = 9.53EE289 pKa = 3.97MAEE292 pKa = 5.71DD293 pKa = 4.13FMQAIKK299 pKa = 9.93QLRR302 pKa = 11.84DD303 pKa = 3.07EE304 pKa = 4.36GLEE307 pKa = 4.12YY308 pKa = 10.56IPDD311 pKa = 3.87YY312 pKa = 9.78YY313 pKa = 10.88EE314 pKa = 4.07KK315 pKa = 10.09WFKK318 pKa = 11.34SPDD321 pKa = 3.54PLTFPNIALIYY332 pKa = 10.09SFSFHH337 pKa = 6.17VGYY340 pKa = 10.48RR341 pKa = 11.84KK342 pKa = 9.09QALSDD347 pKa = 3.44AVYY350 pKa = 10.62DD351 pKa = 4.08QITVTYY357 pKa = 9.29SDD359 pKa = 3.67NVNMEE364 pKa = 3.9MYY366 pKa = 10.18KK367 pKa = 10.24EE368 pKa = 3.87YY369 pKa = 10.86SEE371 pKa = 5.63RR372 pKa = 11.84IEE374 pKa = 4.64NEE376 pKa = 3.1IFTILKK382 pKa = 10.31DD383 pKa = 4.28KK384 pKa = 10.58IIHH387 pKa = 5.46EE388 pKa = 4.44DD389 pKa = 3.24KK390 pKa = 10.86RR391 pKa = 11.84LEE393 pKa = 4.08EE394 pKa = 4.38YY395 pKa = 10.32EE396 pKa = 4.46LSALLSMSSASNGILRR412 pKa = 11.84EE413 pKa = 4.16INFGGQKK420 pKa = 9.72VRR422 pKa = 11.84STKK425 pKa = 10.96KK426 pKa = 9.83NMHH429 pKa = 6.66VIDD432 pKa = 5.48DD433 pKa = 4.54IYY435 pKa = 10.49HH436 pKa = 6.33KK437 pKa = 10.54KK438 pKa = 8.56YY439 pKa = 7.24TTDD442 pKa = 3.2IPPVDD447 pKa = 3.97ARR449 pKa = 11.84NPIPLGRR456 pKa = 11.84RR457 pKa = 11.84DD458 pKa = 3.43VPGRR462 pKa = 11.84RR463 pKa = 11.84TRR465 pKa = 11.84AIFILPYY472 pKa = 9.96QYY474 pKa = 10.79FIAQHH479 pKa = 5.69SFAEE483 pKa = 4.12IMLNYY488 pKa = 10.12AKK490 pKa = 10.44RR491 pKa = 11.84EE492 pKa = 3.84RR493 pKa = 11.84EE494 pKa = 3.69YY495 pKa = 11.49SEE497 pKa = 5.63FYY499 pKa = 10.85SQANQVLSYY508 pKa = 11.54GDD510 pKa = 3.22VTRR513 pKa = 11.84YY514 pKa = 10.16LDD516 pKa = 3.99SNSILCFTDD525 pKa = 3.64VSQWDD530 pKa = 3.74ASQHH534 pKa = 4.09NTKK537 pKa = 10.07VLRR540 pKa = 11.84RR541 pKa = 11.84SIIRR545 pKa = 11.84AMKK548 pKa = 10.01RR549 pKa = 11.84LKK551 pKa = 10.53QLTHH555 pKa = 6.97NINIHH560 pKa = 5.39KK561 pKa = 10.19AINIYY566 pKa = 9.94IQSQEE571 pKa = 4.01NLEE574 pKa = 4.01NSYY577 pKa = 11.5VLIDD581 pKa = 4.13KK582 pKa = 10.65KK583 pKa = 10.76AIQYY587 pKa = 8.39GATASGEE594 pKa = 4.15KK595 pKa = 7.52QTKK598 pKa = 9.23IMNSIANKK606 pKa = 10.46ALIQTVLGKK615 pKa = 11.04LMTDD619 pKa = 3.18YY620 pKa = 11.04TFDD623 pKa = 3.17VKK625 pKa = 10.38MIRR628 pKa = 11.84VDD630 pKa = 3.5GDD632 pKa = 3.41DD633 pKa = 3.45NYY635 pKa = 11.59AIVRR639 pKa = 11.84FPIAITEE646 pKa = 4.16KK647 pKa = 10.46LLSEE651 pKa = 4.6FTSKK655 pKa = 10.44LRR657 pKa = 11.84SYY659 pKa = 10.12YY660 pKa = 10.58SEE662 pKa = 3.99MNVKK666 pKa = 9.88VKK668 pKa = 10.67ALASLTGCEE677 pKa = 3.38IAKK680 pKa = 10.08RR681 pKa = 11.84YY682 pKa = 8.51VAGGMLFFRR691 pKa = 11.84AGVNILHH698 pKa = 6.39HH699 pKa = 6.33EE700 pKa = 4.14KK701 pKa = 10.58RR702 pKa = 11.84NQDD705 pKa = 2.94SAYY708 pKa = 11.06DD709 pKa = 3.75MAATLYY715 pKa = 11.24ANYY718 pKa = 9.6IVNALRR724 pKa = 11.84GLTMSRR730 pKa = 11.84TFILTKK736 pKa = 9.36ICQMTSIKK744 pKa = 9.37ITGTLRR750 pKa = 11.84LFPMKK755 pKa = 10.44SILALNSAFKK765 pKa = 11.19VFDD768 pKa = 3.56EE769 pKa = 4.01VDD771 pKa = 3.45YY772 pKa = 11.26VINYY776 pKa = 7.54PISNLFIQLQRR787 pKa = 11.84KK788 pKa = 8.93LSSIKK793 pKa = 10.48AKK795 pKa = 10.72SKK797 pKa = 10.22IADD800 pKa = 4.2NIAKK804 pKa = 10.1SPQFKK809 pKa = 10.72SYY811 pKa = 11.54VEE813 pKa = 4.31LLNKK817 pKa = 10.34SLTTDD822 pKa = 3.35EE823 pKa = 4.61NPIVSDD829 pKa = 4.71GIRR832 pKa = 11.84LTEE835 pKa = 3.83KK836 pKa = 10.91AKK838 pKa = 10.75LNSYY842 pKa = 10.86APIALEE848 pKa = 3.65KK849 pKa = 10.79RR850 pKa = 11.84RR851 pKa = 11.84DD852 pKa = 3.54QFSIMVSFLQNPTTFKK868 pKa = 11.02SEE870 pKa = 4.21TVVTINDD877 pKa = 3.21VLYY880 pKa = 10.23FISGFIKK887 pKa = 9.86IDD889 pKa = 3.36SSTVLPKK896 pKa = 10.75EE897 pKa = 4.55EE898 pKa = 4.46NNTMPLLPAIIKK910 pKa = 8.17RR911 pKa = 11.84TLSYY915 pKa = 10.75FGLRR919 pKa = 11.84THH921 pKa = 7.59DD922 pKa = 3.77YY923 pKa = 10.17DD924 pKa = 3.94IKK926 pKa = 10.91GSSSTVSKK934 pKa = 10.41IIKK937 pKa = 9.37QYY939 pKa = 10.99SVYY942 pKa = 10.25TPGIEE947 pKa = 3.87EE948 pKa = 4.76LYY950 pKa = 10.55EE951 pKa = 4.01IVNKK955 pKa = 10.66SVDD958 pKa = 3.57TIRR961 pKa = 11.84GYY963 pKa = 10.03FASFNVPKK971 pKa = 10.55ADD973 pKa = 2.96VDD975 pKa = 4.32TYY977 pKa = 11.27ISTQMYY983 pKa = 8.78KK984 pKa = 10.03HH985 pKa = 6.74DD986 pKa = 4.07RR987 pKa = 11.84FKK989 pKa = 10.84ILQAYY994 pKa = 8.83IYY996 pKa = 10.59NLLSVNYY1003 pKa = 10.43GMYY1006 pKa = 10.44QLVDD1010 pKa = 4.04LNSARR1015 pKa = 11.84FFDD1018 pKa = 4.73HH1019 pKa = 7.35VIHH1022 pKa = 6.49TPMAKK1027 pKa = 9.17TPTAVFMIDD1036 pKa = 2.87LALRR1040 pKa = 11.84LKK1042 pKa = 10.38IINHH1046 pKa = 6.7CIEE1049 pKa = 4.21KK1050 pKa = 10.99GEE1052 pKa = 4.39IITVSVHH1059 pKa = 6.42ANKK1062 pKa = 9.54TDD1064 pKa = 3.68YY1065 pKa = 11.32LKK1067 pKa = 10.7LWRR1070 pKa = 11.84MLWNVKK1076 pKa = 7.82TMNSPYY1082 pKa = 10.41SKK1084 pKa = 11.05NSMFDD1089 pKa = 3.17EE1090 pKa = 4.64
MM1 pKa = 7.11AQSIVVDD8 pKa = 3.94GDD10 pKa = 3.44YY11 pKa = 11.3DD12 pKa = 3.97ALASRR17 pKa = 11.84YY18 pKa = 10.17LKK20 pKa = 10.37FVYY23 pKa = 10.25DD24 pKa = 4.32FEE26 pKa = 4.73NVTYY30 pKa = 10.35QNNYY34 pKa = 9.06FATDD38 pKa = 3.29KK39 pKa = 10.54FKK41 pKa = 11.23KK42 pKa = 10.42DD43 pKa = 2.7IEE45 pKa = 4.37QYY47 pKa = 10.67LKK49 pKa = 10.71SIHH52 pKa = 7.03DD53 pKa = 3.97GEE55 pKa = 5.93KK56 pKa = 8.92ITQSKK61 pKa = 9.92IDD63 pKa = 3.87EE64 pKa = 4.38KK65 pKa = 11.34EE66 pKa = 3.93KK67 pKa = 10.74ILLDD71 pKa = 3.42RR72 pKa = 11.84VPAEE76 pKa = 4.13EE77 pKa = 4.14RR78 pKa = 11.84CLISKK83 pKa = 9.67LVFAYY88 pKa = 9.75GKK90 pKa = 9.95HH91 pKa = 5.52GNVEE95 pKa = 4.13NKK97 pKa = 9.43LVKK100 pKa = 10.85YY101 pKa = 9.66GVKK104 pKa = 10.21DD105 pKa = 3.86ALSHH109 pKa = 7.09APQKK113 pKa = 10.15DD114 pKa = 3.2AKK116 pKa = 9.9PYY118 pKa = 8.58EE119 pKa = 4.17NNIITSEE126 pKa = 3.85IFKK129 pKa = 10.54EE130 pKa = 3.99KK131 pKa = 10.8SEE133 pKa = 4.07YY134 pKa = 9.44TDD136 pKa = 3.28IYY138 pKa = 9.73MDD140 pKa = 4.0PSINTSCQSNCQAMMFTISEE160 pKa = 4.2MKK162 pKa = 10.56LNNIKK167 pKa = 10.44NAARR171 pKa = 11.84LEE173 pKa = 4.0KK174 pKa = 10.67LFTIIAATINKK185 pKa = 9.71YY186 pKa = 10.66GMPRR190 pKa = 11.84HH191 pKa = 4.69NTRR194 pKa = 11.84YY195 pKa = 8.54RR196 pKa = 11.84YY197 pKa = 8.92EE198 pKa = 3.9WEE200 pKa = 3.94TMKK203 pKa = 10.97NKK205 pKa = 9.83PYY207 pKa = 10.77HH208 pKa = 5.31LAAWINSSIEE218 pKa = 3.9MIACVVDD225 pKa = 2.54HH226 pKa = 6.41HH227 pKa = 6.96TYY229 pKa = 9.82MIARR233 pKa = 11.84EE234 pKa = 4.43LIVKK238 pKa = 9.94SFTNRR243 pKa = 11.84TSLAKK248 pKa = 10.32LVSSPMTVLTAMLPIRR264 pKa = 11.84GTFITTEE271 pKa = 3.96NLEE274 pKa = 4.19LEE276 pKa = 4.45YY277 pKa = 11.13SNKK280 pKa = 10.12SINYY284 pKa = 8.98LISKK288 pKa = 9.53EE289 pKa = 3.97MAEE292 pKa = 5.71DD293 pKa = 4.13FMQAIKK299 pKa = 9.93QLRR302 pKa = 11.84DD303 pKa = 3.07EE304 pKa = 4.36GLEE307 pKa = 4.12YY308 pKa = 10.56IPDD311 pKa = 3.87YY312 pKa = 9.78YY313 pKa = 10.88EE314 pKa = 4.07KK315 pKa = 10.09WFKK318 pKa = 11.34SPDD321 pKa = 3.54PLTFPNIALIYY332 pKa = 10.09SFSFHH337 pKa = 6.17VGYY340 pKa = 10.48RR341 pKa = 11.84KK342 pKa = 9.09QALSDD347 pKa = 3.44AVYY350 pKa = 10.62DD351 pKa = 4.08QITVTYY357 pKa = 9.29SDD359 pKa = 3.67NVNMEE364 pKa = 3.9MYY366 pKa = 10.18KK367 pKa = 10.24EE368 pKa = 3.87YY369 pKa = 10.86SEE371 pKa = 5.63RR372 pKa = 11.84IEE374 pKa = 4.64NEE376 pKa = 3.1IFTILKK382 pKa = 10.31DD383 pKa = 4.28KK384 pKa = 10.58IIHH387 pKa = 5.46EE388 pKa = 4.44DD389 pKa = 3.24KK390 pKa = 10.86RR391 pKa = 11.84LEE393 pKa = 4.08EE394 pKa = 4.38YY395 pKa = 10.32EE396 pKa = 4.46LSALLSMSSASNGILRR412 pKa = 11.84EE413 pKa = 4.16INFGGQKK420 pKa = 9.72VRR422 pKa = 11.84STKK425 pKa = 10.96KK426 pKa = 9.83NMHH429 pKa = 6.66VIDD432 pKa = 5.48DD433 pKa = 4.54IYY435 pKa = 10.49HH436 pKa = 6.33KK437 pKa = 10.54KK438 pKa = 8.56YY439 pKa = 7.24TTDD442 pKa = 3.2IPPVDD447 pKa = 3.97ARR449 pKa = 11.84NPIPLGRR456 pKa = 11.84RR457 pKa = 11.84DD458 pKa = 3.43VPGRR462 pKa = 11.84RR463 pKa = 11.84TRR465 pKa = 11.84AIFILPYY472 pKa = 9.96QYY474 pKa = 10.79FIAQHH479 pKa = 5.69SFAEE483 pKa = 4.12IMLNYY488 pKa = 10.12AKK490 pKa = 10.44RR491 pKa = 11.84EE492 pKa = 3.84RR493 pKa = 11.84EE494 pKa = 3.69YY495 pKa = 11.49SEE497 pKa = 5.63FYY499 pKa = 10.85SQANQVLSYY508 pKa = 11.54GDD510 pKa = 3.22VTRR513 pKa = 11.84YY514 pKa = 10.16LDD516 pKa = 3.99SNSILCFTDD525 pKa = 3.64VSQWDD530 pKa = 3.74ASQHH534 pKa = 4.09NTKK537 pKa = 10.07VLRR540 pKa = 11.84RR541 pKa = 11.84SIIRR545 pKa = 11.84AMKK548 pKa = 10.01RR549 pKa = 11.84LKK551 pKa = 10.53QLTHH555 pKa = 6.97NINIHH560 pKa = 5.39KK561 pKa = 10.19AINIYY566 pKa = 9.94IQSQEE571 pKa = 4.01NLEE574 pKa = 4.01NSYY577 pKa = 11.5VLIDD581 pKa = 4.13KK582 pKa = 10.65KK583 pKa = 10.76AIQYY587 pKa = 8.39GATASGEE594 pKa = 4.15KK595 pKa = 7.52QTKK598 pKa = 9.23IMNSIANKK606 pKa = 10.46ALIQTVLGKK615 pKa = 11.04LMTDD619 pKa = 3.18YY620 pKa = 11.04TFDD623 pKa = 3.17VKK625 pKa = 10.38MIRR628 pKa = 11.84VDD630 pKa = 3.5GDD632 pKa = 3.41DD633 pKa = 3.45NYY635 pKa = 11.59AIVRR639 pKa = 11.84FPIAITEE646 pKa = 4.16KK647 pKa = 10.46LLSEE651 pKa = 4.6FTSKK655 pKa = 10.44LRR657 pKa = 11.84SYY659 pKa = 10.12YY660 pKa = 10.58SEE662 pKa = 3.99MNVKK666 pKa = 9.88VKK668 pKa = 10.67ALASLTGCEE677 pKa = 3.38IAKK680 pKa = 10.08RR681 pKa = 11.84YY682 pKa = 8.51VAGGMLFFRR691 pKa = 11.84AGVNILHH698 pKa = 6.39HH699 pKa = 6.33EE700 pKa = 4.14KK701 pKa = 10.58RR702 pKa = 11.84NQDD705 pKa = 2.94SAYY708 pKa = 11.06DD709 pKa = 3.75MAATLYY715 pKa = 11.24ANYY718 pKa = 9.6IVNALRR724 pKa = 11.84GLTMSRR730 pKa = 11.84TFILTKK736 pKa = 9.36ICQMTSIKK744 pKa = 9.37ITGTLRR750 pKa = 11.84LFPMKK755 pKa = 10.44SILALNSAFKK765 pKa = 11.19VFDD768 pKa = 3.56EE769 pKa = 4.01VDD771 pKa = 3.45YY772 pKa = 11.26VINYY776 pKa = 7.54PISNLFIQLQRR787 pKa = 11.84KK788 pKa = 8.93LSSIKK793 pKa = 10.48AKK795 pKa = 10.72SKK797 pKa = 10.22IADD800 pKa = 4.2NIAKK804 pKa = 10.1SPQFKK809 pKa = 10.72SYY811 pKa = 11.54VEE813 pKa = 4.31LLNKK817 pKa = 10.34SLTTDD822 pKa = 3.35EE823 pKa = 4.61NPIVSDD829 pKa = 4.71GIRR832 pKa = 11.84LTEE835 pKa = 3.83KK836 pKa = 10.91AKK838 pKa = 10.75LNSYY842 pKa = 10.86APIALEE848 pKa = 3.65KK849 pKa = 10.79RR850 pKa = 11.84RR851 pKa = 11.84DD852 pKa = 3.54QFSIMVSFLQNPTTFKK868 pKa = 11.02SEE870 pKa = 4.21TVVTINDD877 pKa = 3.21VLYY880 pKa = 10.23FISGFIKK887 pKa = 9.86IDD889 pKa = 3.36SSTVLPKK896 pKa = 10.75EE897 pKa = 4.55EE898 pKa = 4.46NNTMPLLPAIIKK910 pKa = 8.17RR911 pKa = 11.84TLSYY915 pKa = 10.75FGLRR919 pKa = 11.84THH921 pKa = 7.59DD922 pKa = 3.77YY923 pKa = 10.17DD924 pKa = 3.94IKK926 pKa = 10.91GSSSTVSKK934 pKa = 10.41IIKK937 pKa = 9.37QYY939 pKa = 10.99SVYY942 pKa = 10.25TPGIEE947 pKa = 3.87EE948 pKa = 4.76LYY950 pKa = 10.55EE951 pKa = 4.01IVNKK955 pKa = 10.66SVDD958 pKa = 3.57TIRR961 pKa = 11.84GYY963 pKa = 10.03FASFNVPKK971 pKa = 10.55ADD973 pKa = 2.96VDD975 pKa = 4.32TYY977 pKa = 11.27ISTQMYY983 pKa = 8.78KK984 pKa = 10.03HH985 pKa = 6.74DD986 pKa = 4.07RR987 pKa = 11.84FKK989 pKa = 10.84ILQAYY994 pKa = 8.83IYY996 pKa = 10.59NLLSVNYY1003 pKa = 10.43GMYY1006 pKa = 10.44QLVDD1010 pKa = 4.04LNSARR1015 pKa = 11.84FFDD1018 pKa = 4.73HH1019 pKa = 7.35VIHH1022 pKa = 6.49TPMAKK1027 pKa = 9.17TPTAVFMIDD1036 pKa = 2.87LALRR1040 pKa = 11.84LKK1042 pKa = 10.38IINHH1046 pKa = 6.7CIEE1049 pKa = 4.21KK1050 pKa = 10.99GEE1052 pKa = 4.39IITVSVHH1059 pKa = 6.42ANKK1062 pKa = 9.54TDD1064 pKa = 3.68YY1065 pKa = 11.32LKK1067 pKa = 10.7LWRR1070 pKa = 11.84MLWNVKK1076 pKa = 7.82TMNSPYY1082 pKa = 10.41SKK1084 pKa = 11.05NSMFDD1089 pKa = 3.17EE1090 pKa = 4.64
Molecular weight: 125.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5608 |
150 |
1090 |
509.8 |
58.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.314 ± 0.369 | 1.409 ± 0.457 |
5.617 ± 0.35 | 5.599 ± 0.514 |
4.297 ± 0.221 | 3.852 ± 0.416 |
1.819 ± 0.215 | 8.827 ± 0.326 |
6.758 ± 0.636 | 8.06 ± 0.383 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.156 ± 0.191 | 7.614 ± 0.446 |
2.889 ± 0.288 | 3.406 ± 0.372 |
4.494 ± 0.191 | 8.006 ± 0.571 |
6.758 ± 0.45 | 5.76 ± 0.287 |
1.088 ± 0.208 | 5.278 ± 0.485 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
