
Fowl aviadenovirus 9
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Aviadenovirus; Fowl aviadenovirus D
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
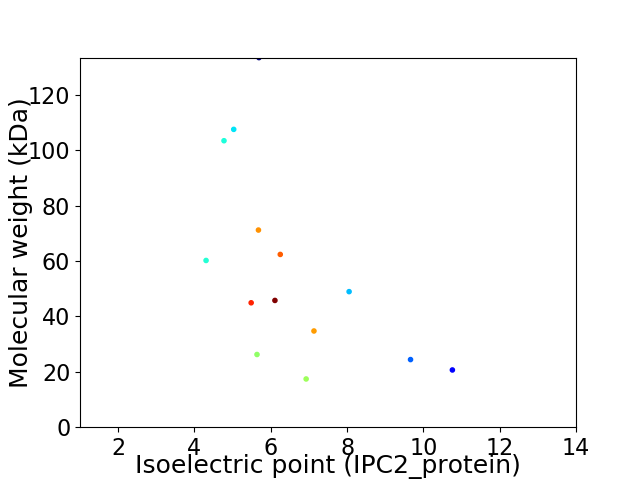
Virtual 2D-PAGE plot for 14 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q77VH6|Q77VH6_9ADEN Pre-hexon-linking protein VIII OS=Fowl aviadenovirus 9 OX=172864 GN=L4 PE=2 SV=1
MM1 pKa = 7.57AKK3 pKa = 8.71STPFAFSMGQHH14 pKa = 6.27SSRR17 pKa = 11.84KK18 pKa = 9.39RR19 pKa = 11.84PADD22 pKa = 3.44SEE24 pKa = 4.19NTQNASKK31 pKa = 10.45VAKK34 pKa = 8.38TQTSATRR41 pKa = 11.84AGVDD45 pKa = 3.89GNDD48 pKa = 4.54DD49 pKa = 4.1LNLVYY54 pKa = 9.94PFWLQNSTSGGGGGGSGGNPSLNPPFIDD82 pKa = 3.73PNGPLYY88 pKa = 10.31VQNSLLYY95 pKa = 10.35VKK97 pKa = 7.76TTAPIEE103 pKa = 4.16VEE105 pKa = 4.18NKK107 pKa = 10.19SLALAYY113 pKa = 10.19DD114 pKa = 3.26SSLAVDD120 pKa = 4.4AQNQLQVKK128 pKa = 9.41VDD130 pKa = 3.49TEE132 pKa = 4.38GPIRR136 pKa = 11.84ISPDD140 pKa = 3.08GLDD143 pKa = 3.18IAVDD147 pKa = 4.11PSTLEE152 pKa = 4.49VDD154 pKa = 4.98DD155 pKa = 4.29EE156 pKa = 4.42WEE158 pKa = 4.15LAVKK162 pKa = 10.47LDD164 pKa = 3.9PNGPLTASSAGININVDD181 pKa = 3.31DD182 pKa = 4.43TLLIEE187 pKa = 6.31DD188 pKa = 5.19DD189 pKa = 4.7DD190 pKa = 5.21ANQAKK195 pKa = 9.98EE196 pKa = 4.27LGVHH200 pKa = 6.76LNPNGPITADD210 pKa = 3.67RR211 pKa = 11.84DD212 pKa = 4.04GLDD215 pKa = 4.84LEE217 pKa = 5.18IDD219 pKa = 3.79SQTMVVKK226 pKa = 10.82DD227 pKa = 3.65SGTSGGVLGVLLKK240 pKa = 10.66PSGGLQSSIQGIGVAVADD258 pKa = 4.23TLTITSNTVEE268 pKa = 4.46VKK270 pKa = 9.86TDD272 pKa = 3.72PNGSISYY279 pKa = 8.85SANGIAVKK287 pKa = 9.85PDD289 pKa = 3.35PSGPLTSSGTGLSVVTAAEE308 pKa = 4.34GSIQSSNAGLAVKK321 pKa = 9.12TDD323 pKa = 3.53PSGPITSGSNGLNLSYY339 pKa = 10.48NASDD343 pKa = 3.55FTVSQGVLNIIRR355 pKa = 11.84NPSTLPDD362 pKa = 3.81AYY364 pKa = 10.91LEE366 pKa = 4.62SGTNYY371 pKa = 10.38LNNFTAQAEE380 pKa = 4.23NSSVFKK386 pKa = 9.71FNCAYY391 pKa = 10.39FLQSWYY397 pKa = 11.27SNGLVTSSLYY407 pKa = 11.01LKK409 pKa = 9.95IDD411 pKa = 3.37RR412 pKa = 11.84AQFSNMPTGQSAEE425 pKa = 3.86NARR428 pKa = 11.84YY429 pKa = 7.46FTFWVPTYY437 pKa = 10.69EE438 pKa = 4.07SLNLSRR444 pKa = 11.84VSTPTITPNTVQWGAFSPAQNCSGNPAFQYY474 pKa = 11.38NLTQPPSIYY483 pKa = 10.33FEE485 pKa = 4.51PKK487 pKa = 9.45SGSVQTFQPVLTGAWNTDD505 pKa = 3.03TYY507 pKa = 11.83NPGTVQVCILPQTVVGGQSTFVNMTCYY534 pKa = 10.52NFRR537 pKa = 11.84CQNPGIFKK545 pKa = 10.21VAASNGTFTIGPIFYY560 pKa = 10.19SCPTNEE566 pKa = 4.2LTRR569 pKa = 11.84PTT571 pKa = 3.88
MM1 pKa = 7.57AKK3 pKa = 8.71STPFAFSMGQHH14 pKa = 6.27SSRR17 pKa = 11.84KK18 pKa = 9.39RR19 pKa = 11.84PADD22 pKa = 3.44SEE24 pKa = 4.19NTQNASKK31 pKa = 10.45VAKK34 pKa = 8.38TQTSATRR41 pKa = 11.84AGVDD45 pKa = 3.89GNDD48 pKa = 4.54DD49 pKa = 4.1LNLVYY54 pKa = 9.94PFWLQNSTSGGGGGGSGGNPSLNPPFIDD82 pKa = 3.73PNGPLYY88 pKa = 10.31VQNSLLYY95 pKa = 10.35VKK97 pKa = 7.76TTAPIEE103 pKa = 4.16VEE105 pKa = 4.18NKK107 pKa = 10.19SLALAYY113 pKa = 10.19DD114 pKa = 3.26SSLAVDD120 pKa = 4.4AQNQLQVKK128 pKa = 9.41VDD130 pKa = 3.49TEE132 pKa = 4.38GPIRR136 pKa = 11.84ISPDD140 pKa = 3.08GLDD143 pKa = 3.18IAVDD147 pKa = 4.11PSTLEE152 pKa = 4.49VDD154 pKa = 4.98DD155 pKa = 4.29EE156 pKa = 4.42WEE158 pKa = 4.15LAVKK162 pKa = 10.47LDD164 pKa = 3.9PNGPLTASSAGININVDD181 pKa = 3.31DD182 pKa = 4.43TLLIEE187 pKa = 6.31DD188 pKa = 5.19DD189 pKa = 4.7DD190 pKa = 5.21ANQAKK195 pKa = 9.98EE196 pKa = 4.27LGVHH200 pKa = 6.76LNPNGPITADD210 pKa = 3.67RR211 pKa = 11.84DD212 pKa = 4.04GLDD215 pKa = 4.84LEE217 pKa = 5.18IDD219 pKa = 3.79SQTMVVKK226 pKa = 10.82DD227 pKa = 3.65SGTSGGVLGVLLKK240 pKa = 10.66PSGGLQSSIQGIGVAVADD258 pKa = 4.23TLTITSNTVEE268 pKa = 4.46VKK270 pKa = 9.86TDD272 pKa = 3.72PNGSISYY279 pKa = 8.85SANGIAVKK287 pKa = 9.85PDD289 pKa = 3.35PSGPLTSSGTGLSVVTAAEE308 pKa = 4.34GSIQSSNAGLAVKK321 pKa = 9.12TDD323 pKa = 3.53PSGPITSGSNGLNLSYY339 pKa = 10.48NASDD343 pKa = 3.55FTVSQGVLNIIRR355 pKa = 11.84NPSTLPDD362 pKa = 3.81AYY364 pKa = 10.91LEE366 pKa = 4.62SGTNYY371 pKa = 10.38LNNFTAQAEE380 pKa = 4.23NSSVFKK386 pKa = 9.71FNCAYY391 pKa = 10.39FLQSWYY397 pKa = 11.27SNGLVTSSLYY407 pKa = 11.01LKK409 pKa = 9.95IDD411 pKa = 3.37RR412 pKa = 11.84AQFSNMPTGQSAEE425 pKa = 3.86NARR428 pKa = 11.84YY429 pKa = 7.46FTFWVPTYY437 pKa = 10.69EE438 pKa = 4.07SLNLSRR444 pKa = 11.84VSTPTITPNTVQWGAFSPAQNCSGNPAFQYY474 pKa = 11.38NLTQPPSIYY483 pKa = 10.33FEE485 pKa = 4.51PKK487 pKa = 9.45SGSVQTFQPVLTGAWNTDD505 pKa = 3.03TYY507 pKa = 11.83NPGTVQVCILPQTVVGGQSTFVNMTCYY534 pKa = 10.52NFRR537 pKa = 11.84CQNPGIFKK545 pKa = 10.21VAASNGTFTIGPIFYY560 pKa = 10.19SCPTNEE566 pKa = 4.2LTRR569 pKa = 11.84PTT571 pKa = 3.88
Molecular weight: 60.24 kDa
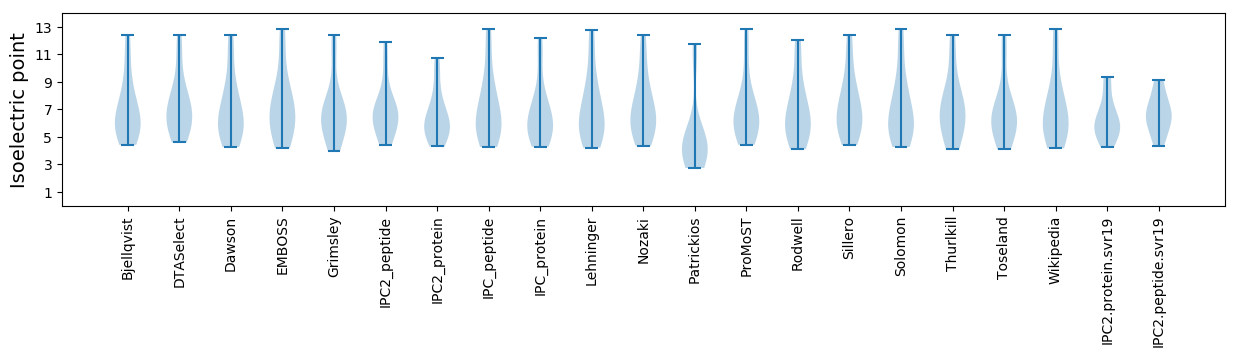
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9QM75|Q9QM75_9ADEN PX OS=Fowl aviadenovirus 9 OX=172864 PE=4 SV=1
MM1 pKa = 8.23DD2 pKa = 4.06YY3 pKa = 11.11AALSPHH9 pKa = 5.83VGGWALRR16 pKa = 11.84DD17 pKa = 3.88GYY19 pKa = 11.25LGDD22 pKa = 3.42SSLRR26 pKa = 11.84GGAINWSNVGSRR38 pKa = 11.84LSSALGTAGRR48 pKa = 11.84WMFNQGNRR56 pKa = 11.84FLNSNTFGQIKK67 pKa = 10.24QGFKK71 pKa = 10.64DD72 pKa = 3.17SGVIRR77 pKa = 11.84NVANLAGEE85 pKa = 4.3TLGALTDD92 pKa = 3.46IGRR95 pKa = 11.84LKK97 pKa = 10.28IQQDD101 pKa = 3.8LEE103 pKa = 4.06KK104 pKa = 10.67LRR106 pKa = 11.84RR107 pKa = 11.84KK108 pKa = 9.96ALGEE112 pKa = 4.63DD113 pKa = 4.23GPATQAEE120 pKa = 4.55LQALIQALQAQVAAGEE136 pKa = 4.64SVPTTAPTVGPSAPAEE152 pKa = 4.31VPRR155 pKa = 11.84PTTRR159 pKa = 11.84PIPEE163 pKa = 4.15MVTEE167 pKa = 3.93VRR169 pKa = 11.84PPVTSSAPAVPEE181 pKa = 3.78TDD183 pKa = 2.94RR184 pKa = 11.84PTTLEE189 pKa = 3.83MRR191 pKa = 11.84PPPAKK196 pKa = 9.84RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 9.85RR201 pKa = 11.84PAPGSWRR208 pKa = 11.84TRR210 pKa = 11.84LNSLSGNGVAVSRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84MCYY228 pKa = 9.89
MM1 pKa = 8.23DD2 pKa = 4.06YY3 pKa = 11.11AALSPHH9 pKa = 5.83VGGWALRR16 pKa = 11.84DD17 pKa = 3.88GYY19 pKa = 11.25LGDD22 pKa = 3.42SSLRR26 pKa = 11.84GGAINWSNVGSRR38 pKa = 11.84LSSALGTAGRR48 pKa = 11.84WMFNQGNRR56 pKa = 11.84FLNSNTFGQIKK67 pKa = 10.24QGFKK71 pKa = 10.64DD72 pKa = 3.17SGVIRR77 pKa = 11.84NVANLAGEE85 pKa = 4.3TLGALTDD92 pKa = 3.46IGRR95 pKa = 11.84LKK97 pKa = 10.28IQQDD101 pKa = 3.8LEE103 pKa = 4.06KK104 pKa = 10.67LRR106 pKa = 11.84RR107 pKa = 11.84KK108 pKa = 9.96ALGEE112 pKa = 4.63DD113 pKa = 4.23GPATQAEE120 pKa = 4.55LQALIQALQAQVAAGEE136 pKa = 4.64SVPTTAPTVGPSAPAEE152 pKa = 4.31VPRR155 pKa = 11.84PTTRR159 pKa = 11.84PIPEE163 pKa = 4.15MVTEE167 pKa = 3.93VRR169 pKa = 11.84PPVTSSAPAVPEE181 pKa = 3.78TDD183 pKa = 2.94RR184 pKa = 11.84PTTLEE189 pKa = 3.83MRR191 pKa = 11.84PPPAKK196 pKa = 9.84RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 9.85RR201 pKa = 11.84PAPGSWRR208 pKa = 11.84TRR210 pKa = 11.84LNSLSGNGVAVSRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84MCYY228 pKa = 9.89
Molecular weight: 24.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7156 |
163 |
1159 |
511.1 |
57.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.622 ± 0.526 | 1.314 ± 0.232 |
6.009 ± 0.368 | 6.358 ± 0.842 |
4.15 ± 0.288 | 6.177 ± 0.483 |
2.194 ± 0.367 | 3.703 ± 0.13 |
3.815 ± 0.39 | 8.147 ± 0.361 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.166 ± 0.24 | 4.919 ± 0.685 |
6.694 ± 0.254 | 4.053 ± 0.291 |
7.183 ± 0.758 | 6.736 ± 0.555 |
6.233 ± 0.395 | 6.47 ± 0.233 |
1.146 ± 0.135 | 3.913 ± 0.438 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
