
Anas platyrhynchos platyrhynchos (Northern mallard)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria; Archelosauria; Archosauria; Dinosauria; Saurischia;
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
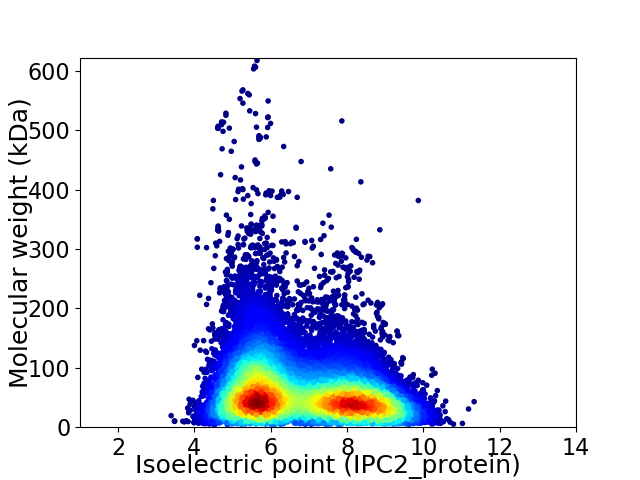
Virtual 2D-PAGE plot for 27053 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U3I454|U3I454_ANAPP Ubiquitin specific peptidase 1 OS=Anas platyrhynchos platyrhynchos OX=8840 GN=USP1 PE=4 SV=2
MM1 pKa = 7.46FDD3 pKa = 3.56CVASEE8 pKa = 4.1FDD10 pKa = 3.41IDD12 pKa = 3.73PARR15 pKa = 11.84TIMVGDD21 pKa = 4.1RR22 pKa = 11.84LDD24 pKa = 3.49TDD26 pKa = 3.12ILMGNTCGLTTLLTLTGVTTLEE48 pKa = 3.95EE49 pKa = 4.41VKK51 pKa = 10.61GHH53 pKa = 6.09EE54 pKa = 4.72EE55 pKa = 3.8SDD57 pKa = 3.78CPARR61 pKa = 11.84QSLVPDD67 pKa = 3.84YY68 pKa = 10.55YY69 pKa = 11.09VDD71 pKa = 4.38SIADD75 pKa = 3.81ILPALQDD82 pKa = 3.29
MM1 pKa = 7.46FDD3 pKa = 3.56CVASEE8 pKa = 4.1FDD10 pKa = 3.41IDD12 pKa = 3.73PARR15 pKa = 11.84TIMVGDD21 pKa = 4.1RR22 pKa = 11.84LDD24 pKa = 3.49TDD26 pKa = 3.12ILMGNTCGLTTLLTLTGVTTLEE48 pKa = 3.95EE49 pKa = 4.41VKK51 pKa = 10.61GHH53 pKa = 6.09EE54 pKa = 4.72EE55 pKa = 3.8SDD57 pKa = 3.78CPARR61 pKa = 11.84QSLVPDD67 pKa = 3.84YY68 pKa = 10.55YY69 pKa = 11.09VDD71 pKa = 4.38SIADD75 pKa = 3.81ILPALQDD82 pKa = 3.29
Molecular weight: 8.94 kDa
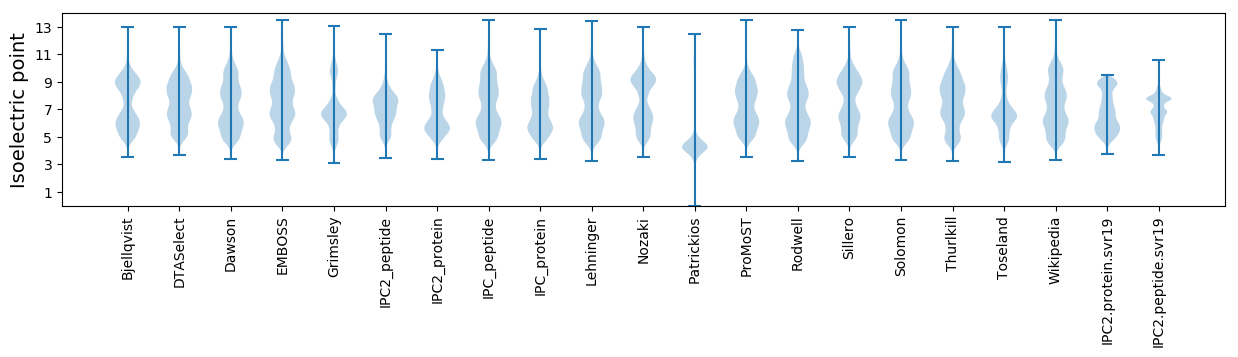
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A493THG4|A0A493THG4_ANAPP Glycine N-acyltransferase-like protein OS=Anas platyrhynchos platyrhynchos OX=8840 GN=GLYATL3 PE=3 SV=1
MM1 pKa = 7.55GFFGQDD7 pKa = 3.14FSVPWGRR14 pKa = 11.84SVPWGHH20 pKa = 6.99PKK22 pKa = 9.28TRR24 pKa = 11.84GHH26 pKa = 6.84PKK28 pKa = 10.45AGGHH32 pKa = 5.65PKK34 pKa = 10.28SRR36 pKa = 11.84GHH38 pKa = 6.57PKK40 pKa = 9.29TRR42 pKa = 11.84GHH44 pKa = 6.74PKK46 pKa = 8.79TRR48 pKa = 11.84RR49 pKa = 11.84HH50 pKa = 5.89PKK52 pKa = 8.87TRR54 pKa = 11.84GHH56 pKa = 6.81PKK58 pKa = 10.5AGGHH62 pKa = 5.7PKK64 pKa = 10.52ARR66 pKa = 11.84GHH68 pKa = 6.48PKK70 pKa = 10.48AGGHH74 pKa = 5.62PKK76 pKa = 10.03PRR78 pKa = 11.84GHH80 pKa = 7.09PKK82 pKa = 9.31TRR84 pKa = 11.84GHH86 pKa = 6.62PNARR90 pKa = 11.84GHH92 pKa = 6.47PKK94 pKa = 10.56AGGHH98 pKa = 6.13PKK100 pKa = 9.3TRR102 pKa = 11.84GHH104 pKa = 6.8PKK106 pKa = 8.63TRR108 pKa = 11.84GYY110 pKa = 9.25PKK112 pKa = 10.24PRR114 pKa = 11.84GHH116 pKa = 6.75PKK118 pKa = 10.26SRR120 pKa = 11.84GHH122 pKa = 6.52PKK124 pKa = 10.45SGGHH128 pKa = 6.31PKK130 pKa = 9.3TRR132 pKa = 11.84GHH134 pKa = 6.94PNTRR138 pKa = 11.84GHH140 pKa = 6.66PKK142 pKa = 9.99PRR144 pKa = 11.84GHH146 pKa = 6.81PKK148 pKa = 10.49ARR150 pKa = 11.84GHH152 pKa = 6.48PKK154 pKa = 10.48AGGHH158 pKa = 5.62PKK160 pKa = 10.07PRR162 pKa = 11.84GHH164 pKa = 6.81PKK166 pKa = 10.46ARR168 pKa = 11.84GHH170 pKa = 6.44PKK172 pKa = 9.27TRR174 pKa = 11.84GHH176 pKa = 6.81PKK178 pKa = 9.23TRR180 pKa = 11.84GHH182 pKa = 6.73PKK184 pKa = 10.38SGGHH188 pKa = 5.86PKK190 pKa = 10.51ARR192 pKa = 11.84GHH194 pKa = 6.39PKK196 pKa = 10.41SGGHH200 pKa = 6.31PKK202 pKa = 9.31TRR204 pKa = 11.84GHH206 pKa = 6.73PKK208 pKa = 10.38SGGHH212 pKa = 5.86PKK214 pKa = 10.5ARR216 pKa = 11.84GHH218 pKa = 6.44PKK220 pKa = 9.27TRR222 pKa = 11.84GHH224 pKa = 6.73PKK226 pKa = 10.34SGGHH230 pKa = 5.85PKK232 pKa = 10.31SRR234 pKa = 11.84GHH236 pKa = 6.44PKK238 pKa = 10.06SGRR241 pKa = 11.84HH242 pKa = 5.01PKK244 pKa = 10.15SRR246 pKa = 11.84GHH248 pKa = 6.23PKK250 pKa = 10.0PRR252 pKa = 11.84GHH254 pKa = 7.07PKK256 pKa = 8.84TRR258 pKa = 11.84RR259 pKa = 11.84HH260 pKa = 6.22PKK262 pKa = 9.49TGGHH266 pKa = 6.4PKK268 pKa = 10.2NRR270 pKa = 11.84GHH272 pKa = 7.06PKK274 pKa = 9.24TRR276 pKa = 11.84GHH278 pKa = 6.61PKK280 pKa = 9.91PRR282 pKa = 11.84GHH284 pKa = 7.09PKK286 pKa = 9.29TRR288 pKa = 11.84GHH290 pKa = 6.74PKK292 pKa = 8.79TRR294 pKa = 11.84RR295 pKa = 11.84HH296 pKa = 5.62PKK298 pKa = 9.35PRR300 pKa = 11.84GHH302 pKa = 6.39RR303 pKa = 11.84KK304 pKa = 9.36SRR306 pKa = 11.84GHH308 pKa = 6.56PKK310 pKa = 9.3TRR312 pKa = 11.84GHH314 pKa = 6.81PKK316 pKa = 9.26TRR318 pKa = 11.84GHH320 pKa = 6.84PKK322 pKa = 10.47AGGHH326 pKa = 6.54PKK328 pKa = 9.98TGGHH332 pKa = 6.73PKK334 pKa = 8.96TRR336 pKa = 11.84APPVPRR342 pKa = 11.84HH343 pKa = 5.43RR344 pKa = 11.84SIPRR348 pKa = 11.84ALPVPRR354 pKa = 11.84PCPGAPWGPPGAAAAAGPGPPLTTPGISRR383 pKa = 11.84SFAPCSPGSRR393 pKa = 11.84RR394 pKa = 11.84RR395 pKa = 11.84RR396 pKa = 11.84RR397 pKa = 11.84RR398 pKa = 11.84NN399 pKa = 2.95
MM1 pKa = 7.55GFFGQDD7 pKa = 3.14FSVPWGRR14 pKa = 11.84SVPWGHH20 pKa = 6.99PKK22 pKa = 9.28TRR24 pKa = 11.84GHH26 pKa = 6.84PKK28 pKa = 10.45AGGHH32 pKa = 5.65PKK34 pKa = 10.28SRR36 pKa = 11.84GHH38 pKa = 6.57PKK40 pKa = 9.29TRR42 pKa = 11.84GHH44 pKa = 6.74PKK46 pKa = 8.79TRR48 pKa = 11.84RR49 pKa = 11.84HH50 pKa = 5.89PKK52 pKa = 8.87TRR54 pKa = 11.84GHH56 pKa = 6.81PKK58 pKa = 10.5AGGHH62 pKa = 5.7PKK64 pKa = 10.52ARR66 pKa = 11.84GHH68 pKa = 6.48PKK70 pKa = 10.48AGGHH74 pKa = 5.62PKK76 pKa = 10.03PRR78 pKa = 11.84GHH80 pKa = 7.09PKK82 pKa = 9.31TRR84 pKa = 11.84GHH86 pKa = 6.62PNARR90 pKa = 11.84GHH92 pKa = 6.47PKK94 pKa = 10.56AGGHH98 pKa = 6.13PKK100 pKa = 9.3TRR102 pKa = 11.84GHH104 pKa = 6.8PKK106 pKa = 8.63TRR108 pKa = 11.84GYY110 pKa = 9.25PKK112 pKa = 10.24PRR114 pKa = 11.84GHH116 pKa = 6.75PKK118 pKa = 10.26SRR120 pKa = 11.84GHH122 pKa = 6.52PKK124 pKa = 10.45SGGHH128 pKa = 6.31PKK130 pKa = 9.3TRR132 pKa = 11.84GHH134 pKa = 6.94PNTRR138 pKa = 11.84GHH140 pKa = 6.66PKK142 pKa = 9.99PRR144 pKa = 11.84GHH146 pKa = 6.81PKK148 pKa = 10.49ARR150 pKa = 11.84GHH152 pKa = 6.48PKK154 pKa = 10.48AGGHH158 pKa = 5.62PKK160 pKa = 10.07PRR162 pKa = 11.84GHH164 pKa = 6.81PKK166 pKa = 10.46ARR168 pKa = 11.84GHH170 pKa = 6.44PKK172 pKa = 9.27TRR174 pKa = 11.84GHH176 pKa = 6.81PKK178 pKa = 9.23TRR180 pKa = 11.84GHH182 pKa = 6.73PKK184 pKa = 10.38SGGHH188 pKa = 5.86PKK190 pKa = 10.51ARR192 pKa = 11.84GHH194 pKa = 6.39PKK196 pKa = 10.41SGGHH200 pKa = 6.31PKK202 pKa = 9.31TRR204 pKa = 11.84GHH206 pKa = 6.73PKK208 pKa = 10.38SGGHH212 pKa = 5.86PKK214 pKa = 10.5ARR216 pKa = 11.84GHH218 pKa = 6.44PKK220 pKa = 9.27TRR222 pKa = 11.84GHH224 pKa = 6.73PKK226 pKa = 10.34SGGHH230 pKa = 5.85PKK232 pKa = 10.31SRR234 pKa = 11.84GHH236 pKa = 6.44PKK238 pKa = 10.06SGRR241 pKa = 11.84HH242 pKa = 5.01PKK244 pKa = 10.15SRR246 pKa = 11.84GHH248 pKa = 6.23PKK250 pKa = 10.0PRR252 pKa = 11.84GHH254 pKa = 7.07PKK256 pKa = 8.84TRR258 pKa = 11.84RR259 pKa = 11.84HH260 pKa = 6.22PKK262 pKa = 9.49TGGHH266 pKa = 6.4PKK268 pKa = 10.2NRR270 pKa = 11.84GHH272 pKa = 7.06PKK274 pKa = 9.24TRR276 pKa = 11.84GHH278 pKa = 6.61PKK280 pKa = 9.91PRR282 pKa = 11.84GHH284 pKa = 7.09PKK286 pKa = 9.29TRR288 pKa = 11.84GHH290 pKa = 6.74PKK292 pKa = 8.79TRR294 pKa = 11.84RR295 pKa = 11.84HH296 pKa = 5.62PKK298 pKa = 9.35PRR300 pKa = 11.84GHH302 pKa = 6.39RR303 pKa = 11.84KK304 pKa = 9.36SRR306 pKa = 11.84GHH308 pKa = 6.56PKK310 pKa = 9.3TRR312 pKa = 11.84GHH314 pKa = 6.81PKK316 pKa = 9.26TRR318 pKa = 11.84GHH320 pKa = 6.84PKK322 pKa = 10.47AGGHH326 pKa = 6.54PKK328 pKa = 9.98TGGHH332 pKa = 6.73PKK334 pKa = 8.96TRR336 pKa = 11.84APPVPRR342 pKa = 11.84HH343 pKa = 5.43RR344 pKa = 11.84SIPRR348 pKa = 11.84ALPVPRR354 pKa = 11.84PCPGAPWGPPGAAAAAGPGPPLTTPGISRR383 pKa = 11.84SFAPCSPGSRR393 pKa = 11.84RR394 pKa = 11.84RR395 pKa = 11.84RR396 pKa = 11.84RR397 pKa = 11.84RR398 pKa = 11.84NN399 pKa = 2.95
Molecular weight: 42.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
16497361 |
36 |
6232 |
609.8 |
67.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.978 ± 0.013 | 2.265 ± 0.011 |
4.866 ± 0.009 | 7.149 ± 0.019 |
3.577 ± 0.01 | 6.428 ± 0.021 |
2.522 ± 0.006 | 4.509 ± 0.011 |
6.021 ± 0.017 | 9.53 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.168 ± 0.006 | 3.905 ± 0.009 |
5.955 ± 0.02 | 4.741 ± 0.012 |
5.579 ± 0.013 | 8.471 ± 0.02 |
5.319 ± 0.011 | 6.074 ± 0.011 |
1.193 ± 0.005 | 2.702 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
