
Tacheng Tick Virus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Chuviridae; Morsusvirus; Morsusvirus argatis
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
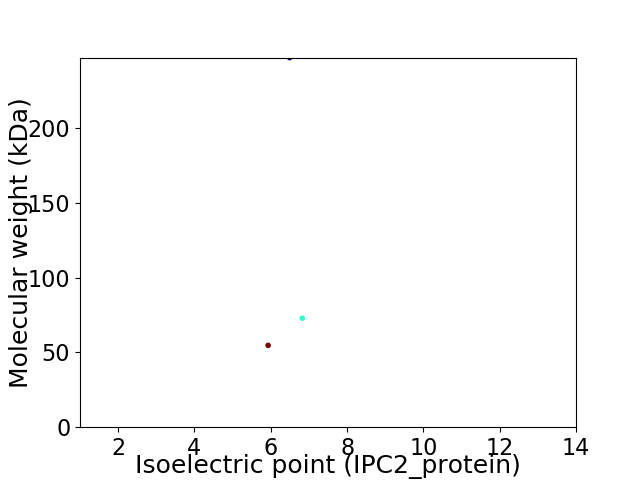
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KT91|A0A0B5KT91_9VIRU GDP polyribonucleotidyltransferase OS=Tacheng Tick Virus 4 OX=1608086 GN=L PE=4 SV=1
MM1 pKa = 7.47TEE3 pKa = 3.35QAAIDD8 pKa = 4.2LRR10 pKa = 11.84DD11 pKa = 3.64HH12 pKa = 6.47PSVYY16 pKa = 10.07RR17 pKa = 11.84RR18 pKa = 11.84TIIVGAEE25 pKa = 3.55EE26 pKa = 3.7RR27 pKa = 11.84AVRR30 pKa = 11.84RR31 pKa = 11.84FATQNPAPGSAPSPQQQYY49 pKa = 11.22KK50 pKa = 10.11DD51 pKa = 3.7LHH53 pKa = 7.86SGDD56 pKa = 4.66LVNADD61 pKa = 3.65TVTRR65 pKa = 11.84LYY67 pKa = 10.03VTDD70 pKa = 3.35TDD72 pKa = 3.57NPEE75 pKa = 3.83RR76 pKa = 11.84NKK78 pKa = 9.94TIRR81 pKa = 11.84VGPMMFEE88 pKa = 4.13RR89 pKa = 11.84NIEE92 pKa = 4.06HH93 pKa = 7.1AFDD96 pKa = 5.05FYY98 pKa = 11.65SEE100 pKa = 4.06GMKK103 pKa = 10.33FAMPRR108 pKa = 11.84ASYY111 pKa = 9.9PPYY114 pKa = 10.79DD115 pKa = 5.0RR116 pKa = 11.84ISSQHH121 pKa = 7.27LEE123 pKa = 4.38IVWAAAVAANMGQEE137 pKa = 4.16VPDD140 pKa = 4.25DD141 pKa = 3.59QFNWMVVGQAFAVGLPDD158 pKa = 3.29HH159 pKa = 6.73VFYY162 pKa = 9.84EE163 pKa = 4.74TIRR166 pKa = 11.84ANEE169 pKa = 3.78IVLAGARR176 pKa = 11.84TMLQKK181 pKa = 9.72MQEE184 pKa = 4.12PARR187 pKa = 11.84TWLLRR192 pKa = 11.84RR193 pKa = 11.84SGNADD198 pKa = 2.96WAVLNSQVEE207 pKa = 4.3MPPEE211 pKa = 5.05GIPMVLEE218 pKa = 4.2ANALHH223 pKa = 6.66LSLGGRR229 pKa = 11.84ARR231 pKa = 11.84MMEE234 pKa = 4.11AYY236 pKa = 9.39MGIFLAYY243 pKa = 7.52TQQGNITVQKK253 pKa = 9.34LDD255 pKa = 4.37KK256 pKa = 10.26IIKK259 pKa = 9.8QIAEE263 pKa = 3.97VQINVALTPANINTLWAKK281 pKa = 10.02FGPFFNEE288 pKa = 3.09EE289 pKa = 3.24AAAYY293 pKa = 7.37FFPRR297 pKa = 11.84WADD300 pKa = 3.57LFADD304 pKa = 3.74RR305 pKa = 11.84HH306 pKa = 5.11IRR308 pKa = 11.84FRR310 pKa = 11.84LIFVQAANAGLTGLTLVRR328 pKa = 11.84DD329 pKa = 4.02MLALYY334 pKa = 9.33PKK336 pKa = 10.17FPWHH340 pKa = 6.66KK341 pKa = 10.35LAAQYY346 pKa = 9.24PAEE349 pKa = 3.9WQAFLKK355 pKa = 10.77ADD357 pKa = 3.57ATVGTNVYY365 pKa = 10.07YY366 pKa = 10.83GYY368 pKa = 9.07TRR370 pKa = 11.84DD371 pKa = 3.55LGIVSVRR378 pKa = 11.84NYY380 pKa = 10.57VPIVYY385 pKa = 9.49CAKK388 pKa = 9.4QLRR391 pKa = 11.84EE392 pKa = 4.21KK393 pKa = 11.07VHH395 pKa = 6.84GDD397 pKa = 3.15QNMQRR402 pKa = 11.84LMTISTFTVQHH413 pKa = 6.39APVIDD418 pKa = 3.74NLVDD422 pKa = 3.31DD423 pKa = 5.19FRR425 pKa = 11.84RR426 pKa = 11.84HH427 pKa = 3.55ITTRR431 pKa = 11.84IGPGDD436 pKa = 3.74RR437 pKa = 11.84EE438 pKa = 4.31LACQYY443 pKa = 7.51YY444 pKa = 6.24TQKK447 pKa = 11.24GEE449 pKa = 4.51AIPEE453 pKa = 4.27GFAMDD458 pKa = 4.88PPAPQPGAGEE468 pKa = 4.01ADD470 pKa = 3.2MVQRR474 pKa = 11.84LVEE477 pKa = 4.06AMRR480 pKa = 11.84GVPQQQ485 pKa = 3.11
MM1 pKa = 7.47TEE3 pKa = 3.35QAAIDD8 pKa = 4.2LRR10 pKa = 11.84DD11 pKa = 3.64HH12 pKa = 6.47PSVYY16 pKa = 10.07RR17 pKa = 11.84RR18 pKa = 11.84TIIVGAEE25 pKa = 3.55EE26 pKa = 3.7RR27 pKa = 11.84AVRR30 pKa = 11.84RR31 pKa = 11.84FATQNPAPGSAPSPQQQYY49 pKa = 11.22KK50 pKa = 10.11DD51 pKa = 3.7LHH53 pKa = 7.86SGDD56 pKa = 4.66LVNADD61 pKa = 3.65TVTRR65 pKa = 11.84LYY67 pKa = 10.03VTDD70 pKa = 3.35TDD72 pKa = 3.57NPEE75 pKa = 3.83RR76 pKa = 11.84NKK78 pKa = 9.94TIRR81 pKa = 11.84VGPMMFEE88 pKa = 4.13RR89 pKa = 11.84NIEE92 pKa = 4.06HH93 pKa = 7.1AFDD96 pKa = 5.05FYY98 pKa = 11.65SEE100 pKa = 4.06GMKK103 pKa = 10.33FAMPRR108 pKa = 11.84ASYY111 pKa = 9.9PPYY114 pKa = 10.79DD115 pKa = 5.0RR116 pKa = 11.84ISSQHH121 pKa = 7.27LEE123 pKa = 4.38IVWAAAVAANMGQEE137 pKa = 4.16VPDD140 pKa = 4.25DD141 pKa = 3.59QFNWMVVGQAFAVGLPDD158 pKa = 3.29HH159 pKa = 6.73VFYY162 pKa = 9.84EE163 pKa = 4.74TIRR166 pKa = 11.84ANEE169 pKa = 3.78IVLAGARR176 pKa = 11.84TMLQKK181 pKa = 9.72MQEE184 pKa = 4.12PARR187 pKa = 11.84TWLLRR192 pKa = 11.84RR193 pKa = 11.84SGNADD198 pKa = 2.96WAVLNSQVEE207 pKa = 4.3MPPEE211 pKa = 5.05GIPMVLEE218 pKa = 4.2ANALHH223 pKa = 6.66LSLGGRR229 pKa = 11.84ARR231 pKa = 11.84MMEE234 pKa = 4.11AYY236 pKa = 9.39MGIFLAYY243 pKa = 7.52TQQGNITVQKK253 pKa = 9.34LDD255 pKa = 4.37KK256 pKa = 10.26IIKK259 pKa = 9.8QIAEE263 pKa = 3.97VQINVALTPANINTLWAKK281 pKa = 10.02FGPFFNEE288 pKa = 3.09EE289 pKa = 3.24AAAYY293 pKa = 7.37FFPRR297 pKa = 11.84WADD300 pKa = 3.57LFADD304 pKa = 3.74RR305 pKa = 11.84HH306 pKa = 5.11IRR308 pKa = 11.84FRR310 pKa = 11.84LIFVQAANAGLTGLTLVRR328 pKa = 11.84DD329 pKa = 4.02MLALYY334 pKa = 9.33PKK336 pKa = 10.17FPWHH340 pKa = 6.66KK341 pKa = 10.35LAAQYY346 pKa = 9.24PAEE349 pKa = 3.9WQAFLKK355 pKa = 10.77ADD357 pKa = 3.57ATVGTNVYY365 pKa = 10.07YY366 pKa = 10.83GYY368 pKa = 9.07TRR370 pKa = 11.84DD371 pKa = 3.55LGIVSVRR378 pKa = 11.84NYY380 pKa = 10.57VPIVYY385 pKa = 9.49CAKK388 pKa = 9.4QLRR391 pKa = 11.84EE392 pKa = 4.21KK393 pKa = 11.07VHH395 pKa = 6.84GDD397 pKa = 3.15QNMQRR402 pKa = 11.84LMTISTFTVQHH413 pKa = 6.39APVIDD418 pKa = 3.74NLVDD422 pKa = 3.31DD423 pKa = 5.19FRR425 pKa = 11.84RR426 pKa = 11.84HH427 pKa = 3.55ITTRR431 pKa = 11.84IGPGDD436 pKa = 3.74RR437 pKa = 11.84EE438 pKa = 4.31LACQYY443 pKa = 7.51YY444 pKa = 6.24TQKK447 pKa = 11.24GEE449 pKa = 4.51AIPEE453 pKa = 4.27GFAMDD458 pKa = 4.88PPAPQPGAGEE468 pKa = 4.01ADD470 pKa = 3.2MVQRR474 pKa = 11.84LVEE477 pKa = 4.06AMRR480 pKa = 11.84GVPQQQ485 pKa = 3.11
Molecular weight: 54.71 kDa
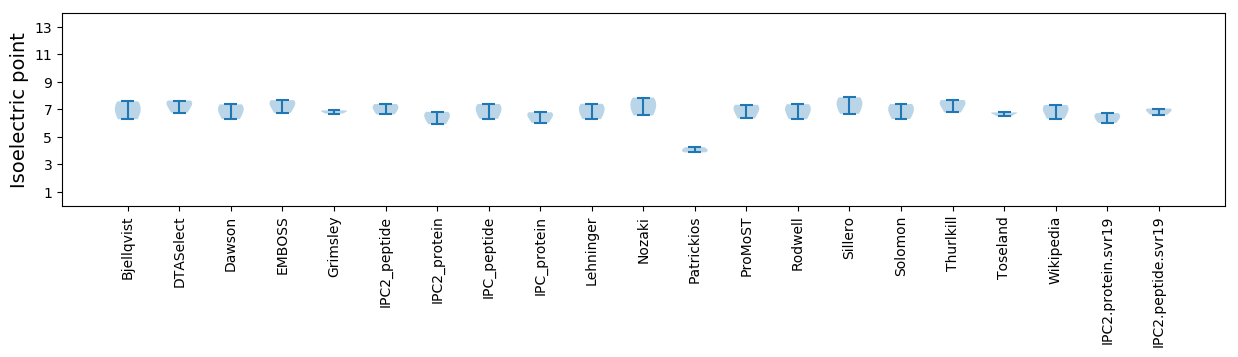
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KRA3|A0A0B5KRA3_9VIRU Putative nucleoprotein OS=Tacheng Tick Virus 4 OX=1608086 GN=N PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84ALCGVGGVLILVVQSIMTAPNTTTRR27 pKa = 11.84GSAVVTAFDD36 pKa = 4.65CNQTTLDD43 pKa = 3.63TVTYY47 pKa = 10.22DD48 pKa = 3.27LTRR51 pKa = 11.84VAEE54 pKa = 4.52CPDD57 pKa = 2.91ISSRR61 pKa = 11.84DD62 pKa = 3.31ISEE65 pKa = 4.15TTRR68 pKa = 11.84LVQVIQQKK76 pKa = 9.76EE77 pKa = 4.13YY78 pKa = 10.64GVLRR82 pKa = 11.84VYY84 pKa = 10.37RR85 pKa = 11.84CHH87 pKa = 5.83VTVTPQIWHH96 pKa = 6.96CGMHH100 pKa = 5.46SHH102 pKa = 6.54TSIVSAPFSAQPLDD116 pKa = 3.77VSEE119 pKa = 4.26QEE121 pKa = 4.18CRR123 pKa = 11.84LMQMDD128 pKa = 4.97GVYY131 pKa = 8.72KK132 pKa = 9.6TATGAIVRR140 pKa = 11.84LKK142 pKa = 10.88RR143 pKa = 11.84NTTSLHH149 pKa = 5.23TVDD152 pKa = 3.89VAGKK156 pKa = 7.29VHH158 pKa = 7.57PDD160 pKa = 3.55GTCVGGSSHH169 pKa = 7.36INGQPYY175 pKa = 8.44YY176 pKa = 10.61SVVHH180 pKa = 6.39RR181 pKa = 11.84EE182 pKa = 3.96AIQIRR187 pKa = 11.84LEE189 pKa = 4.42DD190 pKa = 3.69KK191 pKa = 10.51LVHH194 pKa = 5.8YY195 pKa = 10.18KK196 pKa = 10.23IGDD199 pKa = 3.86TIVVGFGGVTCRR211 pKa = 11.84LADD214 pKa = 3.84ISCHH218 pKa = 6.14DD219 pKa = 3.83FDD221 pKa = 4.48VGRR224 pKa = 11.84MYY226 pKa = 8.22WTLDD230 pKa = 3.18AAGCLDD236 pKa = 3.47GKK238 pKa = 11.21YY239 pKa = 10.42DD240 pKa = 3.77VLYY243 pKa = 10.16TGDD246 pKa = 3.33AAIIHH251 pKa = 5.84TAHH254 pKa = 7.39LGPNKK259 pKa = 10.29EE260 pKa = 3.96YY261 pKa = 10.26MRR263 pKa = 11.84VLRR266 pKa = 11.84RR267 pKa = 11.84ASTPFQIFTPLLGDD281 pKa = 4.14DD282 pKa = 4.03SACGVEE288 pKa = 5.2VRR290 pKa = 11.84TTPYY294 pKa = 10.49AGLYY298 pKa = 8.09VALYY302 pKa = 9.8QSSPPFPIGPITYY315 pKa = 10.38SNVDD319 pKa = 3.23VVKK322 pKa = 10.44YY323 pKa = 10.37LEE325 pKa = 4.19AKK327 pKa = 9.22MVTFAYY333 pKa = 10.2GSALTARR340 pKa = 11.84EE341 pKa = 3.78MVVNLEE347 pKa = 4.02RR348 pKa = 11.84QQCSLHH354 pKa = 7.19RR355 pKa = 11.84EE356 pKa = 3.91TLLNRR361 pKa = 11.84LHH363 pKa = 6.68IVASTPSSATLLLDD377 pKa = 3.54NEE379 pKa = 4.54PGVFGRR385 pKa = 11.84VAGEE389 pKa = 4.04VVHH392 pKa = 6.86LLRR395 pKa = 11.84CPAVEE400 pKa = 4.15VTLRR404 pKa = 11.84RR405 pKa = 11.84ADD407 pKa = 3.46HH408 pKa = 6.94CSLEE412 pKa = 3.8IPIRR416 pKa = 11.84HH417 pKa = 6.91LGRR420 pKa = 11.84DD421 pKa = 3.29VYY423 pKa = 10.35MEE425 pKa = 4.06PRR427 pKa = 11.84TRR429 pKa = 11.84IITTEE434 pKa = 3.89YY435 pKa = 8.59THH437 pKa = 7.04RR438 pKa = 11.84EE439 pKa = 4.22CGPLVPVMFRR449 pKa = 11.84HH450 pKa = 6.18GNQWVQFSPQYY461 pKa = 11.03SPAPEE466 pKa = 4.1PKK468 pKa = 9.36KK469 pKa = 9.75IHH471 pKa = 6.27PMSPTLPSFEE481 pKa = 4.45RR482 pKa = 11.84LVAYY486 pKa = 10.12ARR488 pKa = 11.84GGIYY492 pKa = 10.05DD493 pKa = 4.11PRR495 pKa = 11.84DD496 pKa = 3.33LQQLRR501 pKa = 11.84DD502 pKa = 4.71FILFSHH508 pKa = 6.32KK509 pKa = 9.97RR510 pKa = 11.84DD511 pKa = 4.01AIEE514 pKa = 4.01TNIVAGLSGQSAPKK528 pKa = 10.15YY529 pKa = 10.68DD530 pKa = 3.66LTQVLNSGGWEE541 pKa = 3.67EE542 pKa = 4.05LARR545 pKa = 11.84DD546 pKa = 4.04RR547 pKa = 11.84LQQIWGKK554 pKa = 9.21FQGLGKK560 pKa = 9.91IVNGLVGIYY569 pKa = 9.55VCFVIVRR576 pKa = 11.84ITFGLILRR584 pKa = 11.84TFTIYY589 pKa = 8.91NTHH592 pKa = 6.13GCTKK596 pKa = 10.65LMLGVWSSALTHH608 pKa = 6.4FLLSLAGRR616 pKa = 11.84EE617 pKa = 3.91RR618 pKa = 11.84QEE620 pKa = 3.86PRR622 pKa = 11.84EE623 pKa = 4.03EE624 pKa = 4.12SAGRR628 pKa = 11.84YY629 pKa = 9.65APVDD633 pKa = 3.53QPRR636 pKa = 11.84PPPEE640 pKa = 3.82ATEE643 pKa = 4.66FPSSGQGSIYY653 pKa = 10.09PPIGLL658 pKa = 4.23
MM1 pKa = 7.86RR2 pKa = 11.84ALCGVGGVLILVVQSIMTAPNTTTRR27 pKa = 11.84GSAVVTAFDD36 pKa = 4.65CNQTTLDD43 pKa = 3.63TVTYY47 pKa = 10.22DD48 pKa = 3.27LTRR51 pKa = 11.84VAEE54 pKa = 4.52CPDD57 pKa = 2.91ISSRR61 pKa = 11.84DD62 pKa = 3.31ISEE65 pKa = 4.15TTRR68 pKa = 11.84LVQVIQQKK76 pKa = 9.76EE77 pKa = 4.13YY78 pKa = 10.64GVLRR82 pKa = 11.84VYY84 pKa = 10.37RR85 pKa = 11.84CHH87 pKa = 5.83VTVTPQIWHH96 pKa = 6.96CGMHH100 pKa = 5.46SHH102 pKa = 6.54TSIVSAPFSAQPLDD116 pKa = 3.77VSEE119 pKa = 4.26QEE121 pKa = 4.18CRR123 pKa = 11.84LMQMDD128 pKa = 4.97GVYY131 pKa = 8.72KK132 pKa = 9.6TATGAIVRR140 pKa = 11.84LKK142 pKa = 10.88RR143 pKa = 11.84NTTSLHH149 pKa = 5.23TVDD152 pKa = 3.89VAGKK156 pKa = 7.29VHH158 pKa = 7.57PDD160 pKa = 3.55GTCVGGSSHH169 pKa = 7.36INGQPYY175 pKa = 8.44YY176 pKa = 10.61SVVHH180 pKa = 6.39RR181 pKa = 11.84EE182 pKa = 3.96AIQIRR187 pKa = 11.84LEE189 pKa = 4.42DD190 pKa = 3.69KK191 pKa = 10.51LVHH194 pKa = 5.8YY195 pKa = 10.18KK196 pKa = 10.23IGDD199 pKa = 3.86TIVVGFGGVTCRR211 pKa = 11.84LADD214 pKa = 3.84ISCHH218 pKa = 6.14DD219 pKa = 3.83FDD221 pKa = 4.48VGRR224 pKa = 11.84MYY226 pKa = 8.22WTLDD230 pKa = 3.18AAGCLDD236 pKa = 3.47GKK238 pKa = 11.21YY239 pKa = 10.42DD240 pKa = 3.77VLYY243 pKa = 10.16TGDD246 pKa = 3.33AAIIHH251 pKa = 5.84TAHH254 pKa = 7.39LGPNKK259 pKa = 10.29EE260 pKa = 3.96YY261 pKa = 10.26MRR263 pKa = 11.84VLRR266 pKa = 11.84RR267 pKa = 11.84ASTPFQIFTPLLGDD281 pKa = 4.14DD282 pKa = 4.03SACGVEE288 pKa = 5.2VRR290 pKa = 11.84TTPYY294 pKa = 10.49AGLYY298 pKa = 8.09VALYY302 pKa = 9.8QSSPPFPIGPITYY315 pKa = 10.38SNVDD319 pKa = 3.23VVKK322 pKa = 10.44YY323 pKa = 10.37LEE325 pKa = 4.19AKK327 pKa = 9.22MVTFAYY333 pKa = 10.2GSALTARR340 pKa = 11.84EE341 pKa = 3.78MVVNLEE347 pKa = 4.02RR348 pKa = 11.84QQCSLHH354 pKa = 7.19RR355 pKa = 11.84EE356 pKa = 3.91TLLNRR361 pKa = 11.84LHH363 pKa = 6.68IVASTPSSATLLLDD377 pKa = 3.54NEE379 pKa = 4.54PGVFGRR385 pKa = 11.84VAGEE389 pKa = 4.04VVHH392 pKa = 6.86LLRR395 pKa = 11.84CPAVEE400 pKa = 4.15VTLRR404 pKa = 11.84RR405 pKa = 11.84ADD407 pKa = 3.46HH408 pKa = 6.94CSLEE412 pKa = 3.8IPIRR416 pKa = 11.84HH417 pKa = 6.91LGRR420 pKa = 11.84DD421 pKa = 3.29VYY423 pKa = 10.35MEE425 pKa = 4.06PRR427 pKa = 11.84TRR429 pKa = 11.84IITTEE434 pKa = 3.89YY435 pKa = 8.59THH437 pKa = 7.04RR438 pKa = 11.84EE439 pKa = 4.22CGPLVPVMFRR449 pKa = 11.84HH450 pKa = 6.18GNQWVQFSPQYY461 pKa = 11.03SPAPEE466 pKa = 4.1PKK468 pKa = 9.36KK469 pKa = 9.75IHH471 pKa = 6.27PMSPTLPSFEE481 pKa = 4.45RR482 pKa = 11.84LVAYY486 pKa = 10.12ARR488 pKa = 11.84GGIYY492 pKa = 10.05DD493 pKa = 4.11PRR495 pKa = 11.84DD496 pKa = 3.33LQQLRR501 pKa = 11.84DD502 pKa = 4.71FILFSHH508 pKa = 6.32KK509 pKa = 9.97RR510 pKa = 11.84DD511 pKa = 4.01AIEE514 pKa = 4.01TNIVAGLSGQSAPKK528 pKa = 10.15YY529 pKa = 10.68DD530 pKa = 3.66LTQVLNSGGWEE541 pKa = 3.67EE542 pKa = 4.05LARR545 pKa = 11.84DD546 pKa = 4.04RR547 pKa = 11.84LQQIWGKK554 pKa = 9.21FQGLGKK560 pKa = 9.91IVNGLVGIYY569 pKa = 9.55VCFVIVRR576 pKa = 11.84ITFGLILRR584 pKa = 11.84TFTIYY589 pKa = 8.91NTHH592 pKa = 6.13GCTKK596 pKa = 10.65LMLGVWSSALTHH608 pKa = 6.4FLLSLAGRR616 pKa = 11.84EE617 pKa = 3.91RR618 pKa = 11.84QEE620 pKa = 3.86PRR622 pKa = 11.84EE623 pKa = 4.03EE624 pKa = 4.12SAGRR628 pKa = 11.84YY629 pKa = 9.65APVDD633 pKa = 3.53QPRR636 pKa = 11.84PPPEE640 pKa = 3.82ATEE643 pKa = 4.66FPSSGQGSIYY653 pKa = 10.09PPIGLL658 pKa = 4.23
Molecular weight: 72.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3303 |
485 |
2160 |
1101.0 |
124.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.084 ± 0.922 | 1.605 ± 0.364 |
5.147 ± 0.141 | 5.601 ± 0.348 |
3.724 ± 0.252 | 5.298 ± 1.014 |
3.542 ± 0.35 | 6.418 ± 0.432 |
3.542 ± 0.443 | 10.173 ± 1.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.361 ± 0.436 | 3.149 ± 0.372 |
5.631 ± 0.306 | 4.057 ± 0.604 |
6.903 ± 0.051 | 5.359 ± 0.657 |
6.721 ± 0.398 | 6.842 ± 0.938 |
1.332 ± 0.138 | 4.511 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
