
Torque teno virus 13
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.31
Get precalculated fractions of proteins
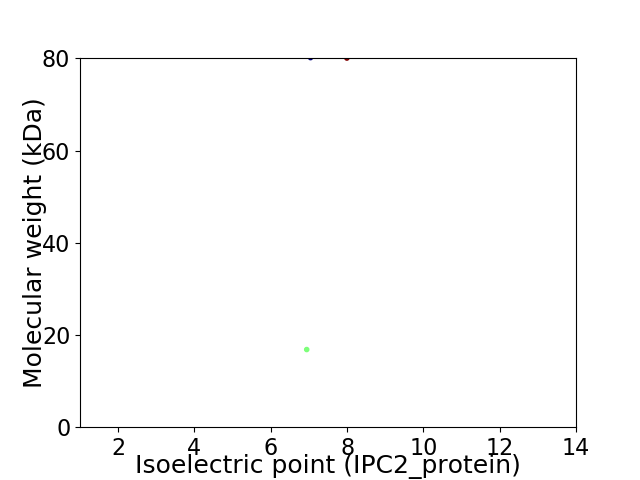
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q99AQ6|Q99AQ6_9VIRU Orf2 OS=Torque teno virus 13 OX=687352 PE=4 SV=1
MM1 pKa = 7.49FLGKK5 pKa = 10.46LYY7 pKa = 10.43RR8 pKa = 11.84KK9 pKa = 9.31KK10 pKa = 10.7RR11 pKa = 11.84KK12 pKa = 9.27LLLQAVRR19 pKa = 11.84APQAPSSMSSSWRR32 pKa = 11.84VPRR35 pKa = 11.84GDD37 pKa = 2.97VSARR41 pKa = 11.84EE42 pKa = 4.17LCWYY46 pKa = 9.96RR47 pKa = 11.84SVRR50 pKa = 11.84EE51 pKa = 4.01SHH53 pKa = 7.55DD54 pKa = 3.66AFCGCRR60 pKa = 11.84DD61 pKa = 3.67PVFHH65 pKa = 7.45LSRR68 pKa = 11.84LAARR72 pKa = 11.84SNHH75 pKa = 5.74QGPPTPPTDD84 pKa = 3.69EE85 pKa = 4.76RR86 pKa = 11.84PSASTPVRR94 pKa = 11.84RR95 pKa = 11.84LLPLPSYY102 pKa = 10.18PGEE105 pKa = 4.49GPQARR110 pKa = 11.84WPGGDD115 pKa = 3.48GEE117 pKa = 4.77GAGDD121 pKa = 3.72ARR123 pKa = 11.84GGAGDD128 pKa = 3.74GGARR132 pKa = 11.84AGEE135 pKa = 4.12EE136 pKa = 4.22EE137 pKa = 4.56YY138 pKa = 10.81RR139 pKa = 11.84PEE141 pKa = 5.46DD142 pKa = 3.65LDD144 pKa = 3.82EE145 pKa = 5.2LFGATEE151 pKa = 4.01QEE153 pKa = 4.2QQ154 pKa = 3.33
MM1 pKa = 7.49FLGKK5 pKa = 10.46LYY7 pKa = 10.43RR8 pKa = 11.84KK9 pKa = 9.31KK10 pKa = 10.7RR11 pKa = 11.84KK12 pKa = 9.27LLLQAVRR19 pKa = 11.84APQAPSSMSSSWRR32 pKa = 11.84VPRR35 pKa = 11.84GDD37 pKa = 2.97VSARR41 pKa = 11.84EE42 pKa = 4.17LCWYY46 pKa = 9.96RR47 pKa = 11.84SVRR50 pKa = 11.84EE51 pKa = 4.01SHH53 pKa = 7.55DD54 pKa = 3.66AFCGCRR60 pKa = 11.84DD61 pKa = 3.67PVFHH65 pKa = 7.45LSRR68 pKa = 11.84LAARR72 pKa = 11.84SNHH75 pKa = 5.74QGPPTPPTDD84 pKa = 3.69EE85 pKa = 4.76RR86 pKa = 11.84PSASTPVRR94 pKa = 11.84RR95 pKa = 11.84LLPLPSYY102 pKa = 10.18PGEE105 pKa = 4.49GPQARR110 pKa = 11.84WPGGDD115 pKa = 3.48GEE117 pKa = 4.77GAGDD121 pKa = 3.72ARR123 pKa = 11.84GGAGDD128 pKa = 3.74GGARR132 pKa = 11.84AGEE135 pKa = 4.12EE136 pKa = 4.22EE137 pKa = 4.56YY138 pKa = 10.81RR139 pKa = 11.84PEE141 pKa = 5.46DD142 pKa = 3.65LDD144 pKa = 3.82EE145 pKa = 5.2LFGATEE151 pKa = 4.01QEE153 pKa = 4.2QQ154 pKa = 3.33
Molecular weight: 16.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q99AQ6|Q99AQ6_9VIRU Orf2 OS=Torque teno virus 13 OX=687352 PE=4 SV=1
MM1 pKa = 7.07PVIWAGMGTGGQNYY15 pKa = 9.48AVRR18 pKa = 11.84SDD20 pKa = 4.01DD21 pKa = 3.71FVVDD25 pKa = 3.65KK26 pKa = 11.52GFGGSFATEE35 pKa = 4.08TFSLRR40 pKa = 11.84VLYY43 pKa = 9.88DD44 pKa = 2.59QHH46 pKa = 7.53QRR48 pKa = 11.84GFNRR52 pKa = 11.84WSHH55 pKa = 5.12TNEE58 pKa = 4.02DD59 pKa = 3.74LDD61 pKa = 3.8LARR64 pKa = 11.84YY65 pKa = 9.11RR66 pKa = 11.84GCKK69 pKa = 4.79WTFYY73 pKa = 10.25RR74 pKa = 11.84HH75 pKa = 6.93PDD77 pKa = 2.97TDD79 pKa = 4.08FIVYY83 pKa = 7.46FTNNPPMKK91 pKa = 8.68TNQYY95 pKa = 7.19TAPLTTPGMLMRR107 pKa = 11.84SKK109 pKa = 10.99YY110 pKa = 10.26KK111 pKa = 10.39ILIPSFKK118 pKa = 10.0TKK120 pKa = 10.41PKK122 pKa = 9.67GKK124 pKa = 8.58KK125 pKa = 8.16TISFRR130 pKa = 11.84ARR132 pKa = 11.84PPKK135 pKa = 10.3LFQDD139 pKa = 3.03KK140 pKa = 9.91WYY142 pKa = 8.12TQQDD146 pKa = 4.26LCPVPLIQLNLTAADD161 pKa = 3.93FTHH164 pKa = 7.36PFGLPLTDD172 pKa = 3.72SPCVRR177 pKa = 11.84FQVLGDD183 pKa = 4.26LYY185 pKa = 11.29NNCLNIDD192 pKa = 4.43LPQFDD197 pKa = 5.0DD198 pKa = 4.48KK199 pKa = 11.54GTISDD204 pKa = 3.53ASSYY208 pKa = 11.45SRR210 pKa = 11.84DD211 pKa = 3.06NKK213 pKa = 9.92QQLEE217 pKa = 4.28EE218 pKa = 4.28LYY220 pKa = 9.13KK221 pKa = 10.4TLFVKK226 pKa = 10.49KK227 pKa = 10.38GCGHH231 pKa = 6.32YY232 pKa = 9.46WQTFMTNSMVKK243 pKa = 10.08AHH245 pKa = 7.05IDD247 pKa = 3.0AAQAQNHH254 pKa = 4.92QQDD257 pKa = 3.43TSGPQSAKK265 pKa = 10.94DD266 pKa = 3.69PFPTKK271 pKa = 9.64PDD273 pKa = 3.49RR274 pKa = 11.84NQFEE278 pKa = 3.85QWKK281 pKa = 10.5NKK283 pKa = 8.23FTDD286 pKa = 3.56PRR288 pKa = 11.84DD289 pKa = 3.91SNFLFATYY297 pKa = 9.82HH298 pKa = 6.63PEE300 pKa = 5.06NITQTIKK307 pKa = 10.13TMRR310 pKa = 11.84DD311 pKa = 2.8NNFALEE317 pKa = 4.1TGKK320 pKa = 10.31NDD322 pKa = 4.71LYY324 pKa = 11.55GDD326 pKa = 3.64YY327 pKa = 10.19QAQYY331 pKa = 9.38TRR333 pKa = 11.84NTHH336 pKa = 6.52LLDD339 pKa = 3.92YY340 pKa = 10.74YY341 pKa = 11.08LGFYY345 pKa = 10.79SPIFLSSGRR354 pKa = 11.84SNTEE358 pKa = 3.05FFTAYY363 pKa = 10.31RR364 pKa = 11.84DD365 pKa = 3.62IIYY368 pKa = 10.85NPLLDD373 pKa = 4.44KK374 pKa = 10.82GTGNMIWFQYY384 pKa = 7.77HH385 pKa = 5.84TKK387 pKa = 9.48TDD389 pKa = 3.86NIFKK393 pKa = 10.47KK394 pKa = 10.22PEE396 pKa = 4.1CHH398 pKa = 6.57WEE400 pKa = 3.99ILDD403 pKa = 3.64MPLWALCNGYY413 pKa = 9.82KK414 pKa = 10.14EE415 pKa = 4.23YY416 pKa = 11.25LEE418 pKa = 4.45SQIKK422 pKa = 10.37YY423 pKa = 10.75GDD425 pKa = 3.41ILVEE429 pKa = 4.09GKK431 pKa = 10.24VLIRR435 pKa = 11.84CPYY438 pKa = 8.16TKK440 pKa = 10.34PPLADD445 pKa = 4.19PNNSLAGYY453 pKa = 8.99VVYY456 pKa = 9.02NTNFGQGKK464 pKa = 8.47WIDD467 pKa = 3.59GKK469 pKa = 11.15GYY471 pKa = 9.7IPLRR475 pKa = 11.84HH476 pKa = 5.78RR477 pKa = 11.84SKK479 pKa = 9.84WYY481 pKa = 10.47VMLMYY486 pKa = 9.71QTDD489 pKa = 3.79VLHH492 pKa = 7.41DD493 pKa = 4.63LVTCGPWQYY502 pKa = 11.46RR503 pKa = 11.84DD504 pKa = 3.79DD505 pKa = 4.26NKK507 pKa = 10.86NSQLIAKK514 pKa = 9.29YY515 pKa = 10.25RR516 pKa = 11.84FTFYY520 pKa = 10.02WGGNMVHH527 pKa = 6.07SQVIRR532 pKa = 11.84NPCKK536 pKa = 9.04DD537 pKa = 3.59TQVSGPRR544 pKa = 11.84RR545 pKa = 11.84QPRR548 pKa = 11.84EE549 pKa = 3.74IQVVDD554 pKa = 3.98PQLITPPWVLHH565 pKa = 6.59SFDD568 pKa = 3.34QRR570 pKa = 11.84RR571 pKa = 11.84GMFTEE576 pKa = 4.12TAIRR580 pKa = 11.84RR581 pKa = 11.84LLRR584 pKa = 11.84QPLPGEE590 pKa = 4.13YY591 pKa = 10.04APPALRR597 pKa = 11.84VPLLFPSSEE606 pKa = 3.95FQRR609 pKa = 11.84EE610 pKa = 4.41GEE612 pKa = 4.23GAEE615 pKa = 4.45SDD617 pKa = 3.59LSSPAKK623 pKa = 10.17RR624 pKa = 11.84PRR626 pKa = 11.84LWQEE630 pKa = 3.72EE631 pKa = 4.08DD632 pKa = 4.7SEE634 pKa = 4.73TQTQSSEE641 pKa = 4.2GPAEE645 pKa = 4.14TTRR648 pKa = 11.84EE649 pKa = 3.86LLEE652 pKa = 4.27RR653 pKa = 11.84KK654 pKa = 9.48LRR656 pKa = 11.84EE657 pKa = 3.84QRR659 pKa = 11.84VLNLQLQQFAVQLAKK674 pKa = 9.61TQANLHH680 pKa = 6.26INPLLYY686 pKa = 10.55SQQQ689 pKa = 3.22
MM1 pKa = 7.07PVIWAGMGTGGQNYY15 pKa = 9.48AVRR18 pKa = 11.84SDD20 pKa = 4.01DD21 pKa = 3.71FVVDD25 pKa = 3.65KK26 pKa = 11.52GFGGSFATEE35 pKa = 4.08TFSLRR40 pKa = 11.84VLYY43 pKa = 9.88DD44 pKa = 2.59QHH46 pKa = 7.53QRR48 pKa = 11.84GFNRR52 pKa = 11.84WSHH55 pKa = 5.12TNEE58 pKa = 4.02DD59 pKa = 3.74LDD61 pKa = 3.8LARR64 pKa = 11.84YY65 pKa = 9.11RR66 pKa = 11.84GCKK69 pKa = 4.79WTFYY73 pKa = 10.25RR74 pKa = 11.84HH75 pKa = 6.93PDD77 pKa = 2.97TDD79 pKa = 4.08FIVYY83 pKa = 7.46FTNNPPMKK91 pKa = 8.68TNQYY95 pKa = 7.19TAPLTTPGMLMRR107 pKa = 11.84SKK109 pKa = 10.99YY110 pKa = 10.26KK111 pKa = 10.39ILIPSFKK118 pKa = 10.0TKK120 pKa = 10.41PKK122 pKa = 9.67GKK124 pKa = 8.58KK125 pKa = 8.16TISFRR130 pKa = 11.84ARR132 pKa = 11.84PPKK135 pKa = 10.3LFQDD139 pKa = 3.03KK140 pKa = 9.91WYY142 pKa = 8.12TQQDD146 pKa = 4.26LCPVPLIQLNLTAADD161 pKa = 3.93FTHH164 pKa = 7.36PFGLPLTDD172 pKa = 3.72SPCVRR177 pKa = 11.84FQVLGDD183 pKa = 4.26LYY185 pKa = 11.29NNCLNIDD192 pKa = 4.43LPQFDD197 pKa = 5.0DD198 pKa = 4.48KK199 pKa = 11.54GTISDD204 pKa = 3.53ASSYY208 pKa = 11.45SRR210 pKa = 11.84DD211 pKa = 3.06NKK213 pKa = 9.92QQLEE217 pKa = 4.28EE218 pKa = 4.28LYY220 pKa = 9.13KK221 pKa = 10.4TLFVKK226 pKa = 10.49KK227 pKa = 10.38GCGHH231 pKa = 6.32YY232 pKa = 9.46WQTFMTNSMVKK243 pKa = 10.08AHH245 pKa = 7.05IDD247 pKa = 3.0AAQAQNHH254 pKa = 4.92QQDD257 pKa = 3.43TSGPQSAKK265 pKa = 10.94DD266 pKa = 3.69PFPTKK271 pKa = 9.64PDD273 pKa = 3.49RR274 pKa = 11.84NQFEE278 pKa = 3.85QWKK281 pKa = 10.5NKK283 pKa = 8.23FTDD286 pKa = 3.56PRR288 pKa = 11.84DD289 pKa = 3.91SNFLFATYY297 pKa = 9.82HH298 pKa = 6.63PEE300 pKa = 5.06NITQTIKK307 pKa = 10.13TMRR310 pKa = 11.84DD311 pKa = 2.8NNFALEE317 pKa = 4.1TGKK320 pKa = 10.31NDD322 pKa = 4.71LYY324 pKa = 11.55GDD326 pKa = 3.64YY327 pKa = 10.19QAQYY331 pKa = 9.38TRR333 pKa = 11.84NTHH336 pKa = 6.52LLDD339 pKa = 3.92YY340 pKa = 10.74YY341 pKa = 11.08LGFYY345 pKa = 10.79SPIFLSSGRR354 pKa = 11.84SNTEE358 pKa = 3.05FFTAYY363 pKa = 10.31RR364 pKa = 11.84DD365 pKa = 3.62IIYY368 pKa = 10.85NPLLDD373 pKa = 4.44KK374 pKa = 10.82GTGNMIWFQYY384 pKa = 7.77HH385 pKa = 5.84TKK387 pKa = 9.48TDD389 pKa = 3.86NIFKK393 pKa = 10.47KK394 pKa = 10.22PEE396 pKa = 4.1CHH398 pKa = 6.57WEE400 pKa = 3.99ILDD403 pKa = 3.64MPLWALCNGYY413 pKa = 9.82KK414 pKa = 10.14EE415 pKa = 4.23YY416 pKa = 11.25LEE418 pKa = 4.45SQIKK422 pKa = 10.37YY423 pKa = 10.75GDD425 pKa = 3.41ILVEE429 pKa = 4.09GKK431 pKa = 10.24VLIRR435 pKa = 11.84CPYY438 pKa = 8.16TKK440 pKa = 10.34PPLADD445 pKa = 4.19PNNSLAGYY453 pKa = 8.99VVYY456 pKa = 9.02NTNFGQGKK464 pKa = 8.47WIDD467 pKa = 3.59GKK469 pKa = 11.15GYY471 pKa = 9.7IPLRR475 pKa = 11.84HH476 pKa = 5.78RR477 pKa = 11.84SKK479 pKa = 9.84WYY481 pKa = 10.47VMLMYY486 pKa = 9.71QTDD489 pKa = 3.79VLHH492 pKa = 7.41DD493 pKa = 4.63LVTCGPWQYY502 pKa = 11.46RR503 pKa = 11.84DD504 pKa = 3.79DD505 pKa = 4.26NKK507 pKa = 10.86NSQLIAKK514 pKa = 9.29YY515 pKa = 10.25RR516 pKa = 11.84FTFYY520 pKa = 10.02WGGNMVHH527 pKa = 6.07SQVIRR532 pKa = 11.84NPCKK536 pKa = 9.04DD537 pKa = 3.59TQVSGPRR544 pKa = 11.84RR545 pKa = 11.84QPRR548 pKa = 11.84EE549 pKa = 3.74IQVVDD554 pKa = 3.98PQLITPPWVLHH565 pKa = 6.59SFDD568 pKa = 3.34QRR570 pKa = 11.84RR571 pKa = 11.84GMFTEE576 pKa = 4.12TAIRR580 pKa = 11.84RR581 pKa = 11.84LLRR584 pKa = 11.84QPLPGEE590 pKa = 4.13YY591 pKa = 10.04APPALRR597 pKa = 11.84VPLLFPSSEE606 pKa = 3.95FQRR609 pKa = 11.84EE610 pKa = 4.41GEE612 pKa = 4.23GAEE615 pKa = 4.45SDD617 pKa = 3.59LSSPAKK623 pKa = 10.17RR624 pKa = 11.84PRR626 pKa = 11.84LWQEE630 pKa = 3.72EE631 pKa = 4.08DD632 pKa = 4.7SEE634 pKa = 4.73TQTQSSEE641 pKa = 4.2GPAEE645 pKa = 4.14TTRR648 pKa = 11.84EE649 pKa = 3.86LLEE652 pKa = 4.27RR653 pKa = 11.84KK654 pKa = 9.48LRR656 pKa = 11.84EE657 pKa = 3.84QRR659 pKa = 11.84VLNLQLQQFAVQLAKK674 pKa = 9.61TQANLHH680 pKa = 6.26INPLLYY686 pKa = 10.55SQQQ689 pKa = 3.22
Molecular weight: 80.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
843 |
154 |
689 |
421.5 |
48.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.457 ± 2.025 | 1.542 ± 0.192 |
6.287 ± 0.209 | 4.745 ± 1.44 |
4.864 ± 1.071 | 6.88 ± 2.273 |
2.254 ± 0.145 | 3.321 ± 1.57 |
5.338 ± 1.295 | 9.015 ± 0.271 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.898 ± 0.283 | 4.508 ± 1.824 |
7.592 ± 1.322 | 6.406 ± 1.186 |
6.88 ± 2.273 | 5.813 ± 1.243 |
6.406 ± 1.8 | 3.915 ± 0.009 |
2.135 ± 0.088 | 4.745 ± 1.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
