
Drosophila melanogaster birnavirus SW-2009a
Taxonomy: Viruses; Riboviria; Orthornavirae; Birnaviridae; unclassified Birnaviridae
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
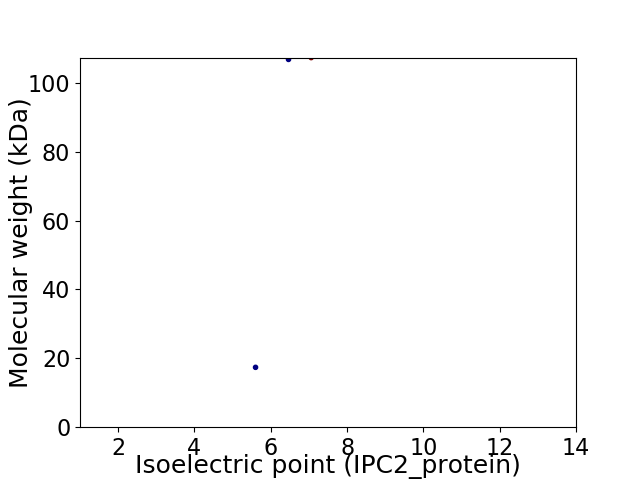
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0U495|D0U495_9VIRU Structural polyprotein OS=Drosophila melanogaster birnavirus SW-2009a OX=663281 PE=4 SV=1
MM1 pKa = 7.96RR2 pKa = 11.84EE3 pKa = 3.67RR4 pKa = 11.84YY5 pKa = 9.55YY6 pKa = 11.03EE7 pKa = 4.05DD8 pKa = 3.7TEE10 pKa = 4.58LNNLSYY16 pKa = 10.96SEE18 pKa = 3.94QDD20 pKa = 3.13KK21 pKa = 10.66QLKK24 pKa = 9.42QICQTSTLDD33 pKa = 3.21QSSCLRR39 pKa = 11.84EE40 pKa = 4.07VPPVYY45 pKa = 9.42QTTTSAVIAYY55 pKa = 7.86DD56 pKa = 3.49KK57 pKa = 10.78KK58 pKa = 10.79QSLRR62 pKa = 11.84TLWSDD67 pKa = 3.73LLEE70 pKa = 4.75KK71 pKa = 11.05GPLFCSLTILAALLALILSMMTRR94 pKa = 11.84ANLTNTPNHH103 pKa = 6.2SLSPNASMSPIITEE117 pKa = 4.01EE118 pKa = 4.22SPQEE122 pKa = 4.35SYY124 pKa = 10.77TSEE127 pKa = 4.26VPHH130 pKa = 6.06SHH132 pKa = 6.12QEE134 pKa = 3.96STTSQEE140 pKa = 3.59QSMPPRR146 pKa = 11.84MKK148 pKa = 10.53AHH150 pKa = 7.13LRR152 pKa = 11.84RR153 pKa = 3.99
MM1 pKa = 7.96RR2 pKa = 11.84EE3 pKa = 3.67RR4 pKa = 11.84YY5 pKa = 9.55YY6 pKa = 11.03EE7 pKa = 4.05DD8 pKa = 3.7TEE10 pKa = 4.58LNNLSYY16 pKa = 10.96SEE18 pKa = 3.94QDD20 pKa = 3.13KK21 pKa = 10.66QLKK24 pKa = 9.42QICQTSTLDD33 pKa = 3.21QSSCLRR39 pKa = 11.84EE40 pKa = 4.07VPPVYY45 pKa = 9.42QTTTSAVIAYY55 pKa = 7.86DD56 pKa = 3.49KK57 pKa = 10.78KK58 pKa = 10.79QSLRR62 pKa = 11.84TLWSDD67 pKa = 3.73LLEE70 pKa = 4.75KK71 pKa = 11.05GPLFCSLTILAALLALILSMMTRR94 pKa = 11.84ANLTNTPNHH103 pKa = 6.2SLSPNASMSPIITEE117 pKa = 4.01EE118 pKa = 4.22SPQEE122 pKa = 4.35SYY124 pKa = 10.77TSEE127 pKa = 4.26VPHH130 pKa = 6.06SHH132 pKa = 6.12QEE134 pKa = 3.96STTSQEE140 pKa = 3.59QSMPPRR146 pKa = 11.84MKK148 pKa = 10.53AHH150 pKa = 7.13LRR152 pKa = 11.84RR153 pKa = 3.99
Molecular weight: 17.38 kDa
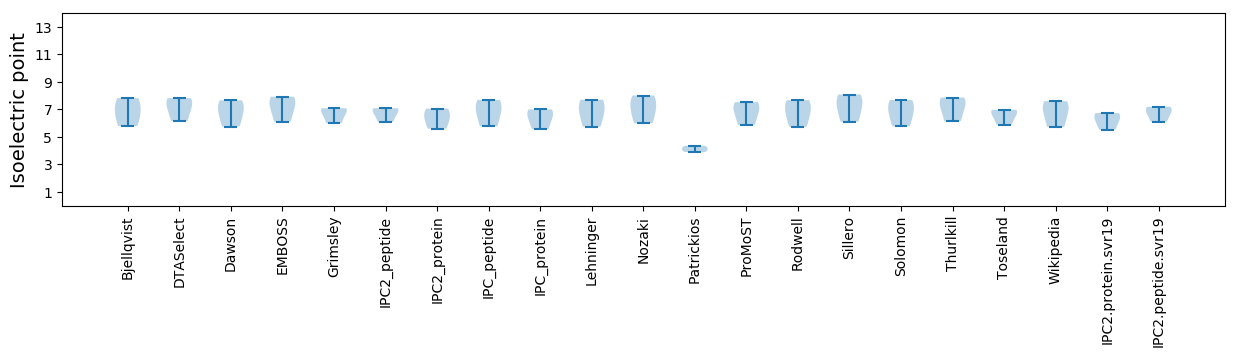
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0U496|D0U496_9VIRU Protein VP1 OS=Drosophila melanogaster birnavirus SW-2009a OX=663281 PE=4 SV=1
MM1 pKa = 7.69NEE3 pKa = 3.92TKK5 pKa = 10.42LVNSTRR11 pKa = 11.84VLEE14 pKa = 4.07IAQYY18 pKa = 8.01IAKK21 pKa = 10.01RR22 pKa = 11.84RR23 pKa = 11.84IHH25 pKa = 6.53AFKK28 pKa = 10.32PYY30 pKa = 10.47YY31 pKa = 10.15LRR33 pKa = 11.84QNLEE37 pKa = 4.09FEE39 pKa = 4.52ASSVLDD45 pKa = 3.91EE46 pKa = 4.32QIVRR50 pKa = 11.84YY51 pKa = 8.78LQTIRR56 pKa = 11.84TLQLEE61 pKa = 4.28SGNPKK66 pKa = 10.46DD67 pKa = 5.62YY68 pKa = 11.22EE69 pKa = 4.51DD70 pKa = 3.78CTKK73 pKa = 10.53IHH75 pKa = 6.8LPDD78 pKa = 5.03GDD80 pKa = 4.17PKK82 pKa = 10.83IPFYY86 pKa = 10.78RR87 pKa = 11.84PRR89 pKa = 11.84FTTGTYY95 pKa = 10.44DD96 pKa = 4.71DD97 pKa = 4.78KK98 pKa = 11.44LLNAIAFQWLASKK111 pKa = 7.7EE112 pKa = 4.41TQGNFEE118 pKa = 4.06TAKK121 pKa = 10.38GYY123 pKa = 9.83VVQRR127 pKa = 11.84DD128 pKa = 4.41RR129 pKa = 11.84IADD132 pKa = 3.78PTDD135 pKa = 2.9MDD137 pKa = 3.97TTIKK141 pKa = 10.35RR142 pKa = 11.84CRR144 pKa = 11.84QACIDD149 pKa = 3.69YY150 pKa = 10.08RR151 pKa = 11.84LSSGTWQGQLARR163 pKa = 11.84LKK165 pKa = 10.62DD166 pKa = 3.69MQNIRR171 pKa = 11.84HH172 pKa = 5.5GVDD175 pKa = 3.24GKK177 pKa = 10.52FLPKK181 pKa = 9.96SLKK184 pKa = 9.95EE185 pKa = 3.8LGYY188 pKa = 10.82DD189 pKa = 3.54LEE191 pKa = 4.54SFSKK195 pKa = 10.42ILEE198 pKa = 3.92KK199 pKa = 10.14TLPINAVDD207 pKa = 4.35LSQTISLRR215 pKa = 11.84YY216 pKa = 9.74LPTLFIEE223 pKa = 4.38EE224 pKa = 4.46TEE226 pKa = 4.36VEE228 pKa = 4.39VKK230 pKa = 10.33GVKK233 pKa = 8.41KK234 pKa = 9.28TSYY237 pKa = 10.55IDD239 pKa = 3.31NTPKK243 pKa = 10.78LKK245 pKa = 10.01PDD247 pKa = 3.64SKK249 pKa = 10.91AGPPFQSKK257 pKa = 8.22YY258 pKa = 10.26RR259 pKa = 11.84KK260 pKa = 9.66GDD262 pKa = 3.37MFSSILALADD272 pKa = 3.82DD273 pKa = 4.77ALLGLSEE280 pKa = 4.58VIHH283 pKa = 6.82GRR285 pKa = 11.84MDD287 pKa = 2.96RR288 pKa = 11.84HH289 pKa = 6.5KK290 pKa = 9.81YY291 pKa = 9.63FKK293 pKa = 10.77DD294 pKa = 3.39RR295 pKa = 11.84IYY297 pKa = 11.27LAMCFLFPKK306 pKa = 9.17EE307 pKa = 3.98EE308 pKa = 4.09RR309 pKa = 11.84YY310 pKa = 9.24EE311 pKa = 4.0ASKK314 pKa = 9.56LTTKK318 pKa = 9.24TRR320 pKa = 11.84NIASVCGVTHH330 pKa = 6.31YY331 pKa = 11.69LMLHH335 pKa = 5.33AMEE338 pKa = 4.88PVMEE342 pKa = 4.18KK343 pKa = 10.76SKK345 pKa = 9.61WNCMSSEE352 pKa = 4.29TYY354 pKa = 10.42EE355 pKa = 4.12NPSLSKK361 pKa = 8.81FTPYY365 pKa = 10.34FGNLQKK371 pKa = 10.86FLDD374 pKa = 4.24FGLHH378 pKa = 5.52RR379 pKa = 11.84EE380 pKa = 4.04KK381 pKa = 10.93KK382 pKa = 9.57KK383 pKa = 11.04VFMPRR388 pKa = 11.84GAKK391 pKa = 7.31PTHH394 pKa = 6.81RR395 pKa = 11.84DD396 pKa = 3.87FIYY399 pKa = 10.71ADD401 pKa = 2.81NWYY404 pKa = 9.85IFYY407 pKa = 10.6RR408 pKa = 11.84NEE410 pKa = 4.88DD411 pKa = 3.47GTTDD415 pKa = 3.84YY416 pKa = 11.68YY417 pKa = 11.72SLDD420 pKa = 3.82LEE422 pKa = 4.53KK423 pKa = 11.37GEE425 pKa = 5.34ANATPDD431 pKa = 2.94IAQCISYY438 pKa = 10.82YY439 pKa = 10.73LLTRR443 pKa = 11.84GWISDD448 pKa = 3.59DD449 pKa = 4.29GIVGFGPTWSTFLTQIVPACIVDD472 pKa = 4.19NISVYY477 pKa = 11.46YY478 pKa = 10.73NMQLRR483 pKa = 11.84FPGQASGNTYY493 pKa = 9.32TFLINHH499 pKa = 7.11AVTTKK504 pKa = 10.42LRR506 pKa = 11.84QEE508 pKa = 4.05WLNRR512 pKa = 11.84GCPRR516 pKa = 11.84PGSAEE521 pKa = 3.67FTQCCKK527 pKa = 9.98KK528 pKa = 9.46TGVNTKK534 pKa = 9.91IEE536 pKa = 4.14FHH538 pKa = 6.53SEE540 pKa = 3.56NLEE543 pKa = 4.03QQVKK547 pKa = 7.53EE548 pKa = 4.69TIANTPNVGFLQGDD562 pKa = 3.78STDD565 pKa = 3.79SVDD568 pKa = 5.41ANGQSWFQKK577 pKa = 10.31ALPRR581 pKa = 11.84LKK583 pKa = 10.55LDD585 pKa = 3.56LLGYY589 pKa = 10.03DD590 pKa = 3.66AVYY593 pKa = 10.72SHH595 pKa = 6.4QLGIFIPVLAEE606 pKa = 3.49EE607 pKa = 4.35RR608 pKa = 11.84FYY610 pKa = 10.92PSMMWPRR617 pKa = 11.84KK618 pKa = 9.58GGEE621 pKa = 4.11TIKK624 pKa = 10.89DD625 pKa = 3.49YY626 pKa = 10.9IQRR629 pKa = 11.84NLYY632 pKa = 9.72NLVRR636 pKa = 11.84YY637 pKa = 8.12EE638 pKa = 3.98VLRR641 pKa = 11.84IMGGWTIPLADD652 pKa = 3.54QALHH656 pKa = 6.58NMATNIRR663 pKa = 11.84TNIVDD668 pKa = 4.12KK669 pKa = 10.26RR670 pKa = 11.84HH671 pKa = 6.29KK672 pKa = 9.89MDD674 pKa = 2.9TSTFLGQEE682 pKa = 4.04DD683 pKa = 4.86LAPDD687 pKa = 3.81LWGAMEE693 pKa = 5.26DD694 pKa = 3.55SGLTTEE700 pKa = 5.24LALNPEE706 pKa = 4.64FSMTHH711 pKa = 5.57EE712 pKa = 4.42LLTRR716 pKa = 11.84LNTPKK721 pKa = 9.08QTSPEE726 pKa = 3.94HH727 pKa = 6.1MPVKK731 pKa = 10.12PKK733 pKa = 9.86PRR735 pKa = 11.84HH736 pKa = 4.71LSKK739 pKa = 10.91PEE741 pKa = 3.77LMADD745 pKa = 3.5LQEE748 pKa = 4.51LKK750 pKa = 10.77KK751 pKa = 11.03SLTNLTSSHH760 pKa = 5.29TEE762 pKa = 3.48KK763 pKa = 11.08LLAKK767 pKa = 9.86PEE769 pKa = 4.09YY770 pKa = 10.07VRR772 pKa = 11.84SKK774 pKa = 10.76LEE776 pKa = 3.89EE777 pKa = 4.1VIKK780 pKa = 9.49TFPSLEE786 pKa = 4.26AIITIMGKK794 pKa = 9.98YY795 pKa = 9.07EE796 pKa = 4.03DD797 pKa = 5.18LYY799 pKa = 11.51SEE801 pKa = 4.75LDD803 pKa = 3.25EE804 pKa = 4.83VYY806 pKa = 10.58NNKK809 pKa = 9.0MKK811 pKa = 10.57IEE813 pKa = 4.28DD814 pKa = 3.86FMDD817 pKa = 5.25KK818 pKa = 9.45LTHH821 pKa = 7.26WINQAQHH828 pKa = 5.33EE829 pKa = 4.66MEE831 pKa = 3.84EE832 pKa = 4.25AMRR835 pKa = 11.84KK836 pKa = 9.33SIRR839 pKa = 11.84VVQQAQVQDD848 pKa = 3.93YY849 pKa = 9.93EE850 pKa = 4.52NIEE853 pKa = 3.88AFYY856 pKa = 10.93EE857 pKa = 4.1KK858 pKa = 10.65DD859 pKa = 3.24PTKK862 pKa = 10.7LADD865 pKa = 3.53PLGYY869 pKa = 10.26KK870 pKa = 10.15GAQTLVGKK878 pKa = 8.49HH879 pKa = 5.41TFPEE883 pKa = 4.43SSSTIPASLTKK894 pKa = 10.28SQKK897 pKa = 10.57KK898 pKa = 9.08NLKK901 pKa = 9.19KK902 pKa = 10.14KK903 pKa = 10.08QRR905 pKa = 11.84ISKK908 pKa = 10.14LRR910 pKa = 11.84DD911 pKa = 2.81IEE913 pKa = 4.14QASWSRR919 pKa = 11.84DD920 pKa = 3.16ALGEE924 pKa = 4.0IRR926 pKa = 11.84KK927 pKa = 9.41HH928 pKa = 6.43EE929 pKa = 4.23EE930 pKa = 3.74DD931 pKa = 3.5LL932 pKa = 4.52
MM1 pKa = 7.69NEE3 pKa = 3.92TKK5 pKa = 10.42LVNSTRR11 pKa = 11.84VLEE14 pKa = 4.07IAQYY18 pKa = 8.01IAKK21 pKa = 10.01RR22 pKa = 11.84RR23 pKa = 11.84IHH25 pKa = 6.53AFKK28 pKa = 10.32PYY30 pKa = 10.47YY31 pKa = 10.15LRR33 pKa = 11.84QNLEE37 pKa = 4.09FEE39 pKa = 4.52ASSVLDD45 pKa = 3.91EE46 pKa = 4.32QIVRR50 pKa = 11.84YY51 pKa = 8.78LQTIRR56 pKa = 11.84TLQLEE61 pKa = 4.28SGNPKK66 pKa = 10.46DD67 pKa = 5.62YY68 pKa = 11.22EE69 pKa = 4.51DD70 pKa = 3.78CTKK73 pKa = 10.53IHH75 pKa = 6.8LPDD78 pKa = 5.03GDD80 pKa = 4.17PKK82 pKa = 10.83IPFYY86 pKa = 10.78RR87 pKa = 11.84PRR89 pKa = 11.84FTTGTYY95 pKa = 10.44DD96 pKa = 4.71DD97 pKa = 4.78KK98 pKa = 11.44LLNAIAFQWLASKK111 pKa = 7.7EE112 pKa = 4.41TQGNFEE118 pKa = 4.06TAKK121 pKa = 10.38GYY123 pKa = 9.83VVQRR127 pKa = 11.84DD128 pKa = 4.41RR129 pKa = 11.84IADD132 pKa = 3.78PTDD135 pKa = 2.9MDD137 pKa = 3.97TTIKK141 pKa = 10.35RR142 pKa = 11.84CRR144 pKa = 11.84QACIDD149 pKa = 3.69YY150 pKa = 10.08RR151 pKa = 11.84LSSGTWQGQLARR163 pKa = 11.84LKK165 pKa = 10.62DD166 pKa = 3.69MQNIRR171 pKa = 11.84HH172 pKa = 5.5GVDD175 pKa = 3.24GKK177 pKa = 10.52FLPKK181 pKa = 9.96SLKK184 pKa = 9.95EE185 pKa = 3.8LGYY188 pKa = 10.82DD189 pKa = 3.54LEE191 pKa = 4.54SFSKK195 pKa = 10.42ILEE198 pKa = 3.92KK199 pKa = 10.14TLPINAVDD207 pKa = 4.35LSQTISLRR215 pKa = 11.84YY216 pKa = 9.74LPTLFIEE223 pKa = 4.38EE224 pKa = 4.46TEE226 pKa = 4.36VEE228 pKa = 4.39VKK230 pKa = 10.33GVKK233 pKa = 8.41KK234 pKa = 9.28TSYY237 pKa = 10.55IDD239 pKa = 3.31NTPKK243 pKa = 10.78LKK245 pKa = 10.01PDD247 pKa = 3.64SKK249 pKa = 10.91AGPPFQSKK257 pKa = 8.22YY258 pKa = 10.26RR259 pKa = 11.84KK260 pKa = 9.66GDD262 pKa = 3.37MFSSILALADD272 pKa = 3.82DD273 pKa = 4.77ALLGLSEE280 pKa = 4.58VIHH283 pKa = 6.82GRR285 pKa = 11.84MDD287 pKa = 2.96RR288 pKa = 11.84HH289 pKa = 6.5KK290 pKa = 9.81YY291 pKa = 9.63FKK293 pKa = 10.77DD294 pKa = 3.39RR295 pKa = 11.84IYY297 pKa = 11.27LAMCFLFPKK306 pKa = 9.17EE307 pKa = 3.98EE308 pKa = 4.09RR309 pKa = 11.84YY310 pKa = 9.24EE311 pKa = 4.0ASKK314 pKa = 9.56LTTKK318 pKa = 9.24TRR320 pKa = 11.84NIASVCGVTHH330 pKa = 6.31YY331 pKa = 11.69LMLHH335 pKa = 5.33AMEE338 pKa = 4.88PVMEE342 pKa = 4.18KK343 pKa = 10.76SKK345 pKa = 9.61WNCMSSEE352 pKa = 4.29TYY354 pKa = 10.42EE355 pKa = 4.12NPSLSKK361 pKa = 8.81FTPYY365 pKa = 10.34FGNLQKK371 pKa = 10.86FLDD374 pKa = 4.24FGLHH378 pKa = 5.52RR379 pKa = 11.84EE380 pKa = 4.04KK381 pKa = 10.93KK382 pKa = 9.57KK383 pKa = 11.04VFMPRR388 pKa = 11.84GAKK391 pKa = 7.31PTHH394 pKa = 6.81RR395 pKa = 11.84DD396 pKa = 3.87FIYY399 pKa = 10.71ADD401 pKa = 2.81NWYY404 pKa = 9.85IFYY407 pKa = 10.6RR408 pKa = 11.84NEE410 pKa = 4.88DD411 pKa = 3.47GTTDD415 pKa = 3.84YY416 pKa = 11.68YY417 pKa = 11.72SLDD420 pKa = 3.82LEE422 pKa = 4.53KK423 pKa = 11.37GEE425 pKa = 5.34ANATPDD431 pKa = 2.94IAQCISYY438 pKa = 10.82YY439 pKa = 10.73LLTRR443 pKa = 11.84GWISDD448 pKa = 3.59DD449 pKa = 4.29GIVGFGPTWSTFLTQIVPACIVDD472 pKa = 4.19NISVYY477 pKa = 11.46YY478 pKa = 10.73NMQLRR483 pKa = 11.84FPGQASGNTYY493 pKa = 9.32TFLINHH499 pKa = 7.11AVTTKK504 pKa = 10.42LRR506 pKa = 11.84QEE508 pKa = 4.05WLNRR512 pKa = 11.84GCPRR516 pKa = 11.84PGSAEE521 pKa = 3.67FTQCCKK527 pKa = 9.98KK528 pKa = 9.46TGVNTKK534 pKa = 9.91IEE536 pKa = 4.14FHH538 pKa = 6.53SEE540 pKa = 3.56NLEE543 pKa = 4.03QQVKK547 pKa = 7.53EE548 pKa = 4.69TIANTPNVGFLQGDD562 pKa = 3.78STDD565 pKa = 3.79SVDD568 pKa = 5.41ANGQSWFQKK577 pKa = 10.31ALPRR581 pKa = 11.84LKK583 pKa = 10.55LDD585 pKa = 3.56LLGYY589 pKa = 10.03DD590 pKa = 3.66AVYY593 pKa = 10.72SHH595 pKa = 6.4QLGIFIPVLAEE606 pKa = 3.49EE607 pKa = 4.35RR608 pKa = 11.84FYY610 pKa = 10.92PSMMWPRR617 pKa = 11.84KK618 pKa = 9.58GGEE621 pKa = 4.11TIKK624 pKa = 10.89DD625 pKa = 3.49YY626 pKa = 10.9IQRR629 pKa = 11.84NLYY632 pKa = 9.72NLVRR636 pKa = 11.84YY637 pKa = 8.12EE638 pKa = 3.98VLRR641 pKa = 11.84IMGGWTIPLADD652 pKa = 3.54QALHH656 pKa = 6.58NMATNIRR663 pKa = 11.84TNIVDD668 pKa = 4.12KK669 pKa = 10.26RR670 pKa = 11.84HH671 pKa = 6.29KK672 pKa = 9.89MDD674 pKa = 2.9TSTFLGQEE682 pKa = 4.04DD683 pKa = 4.86LAPDD687 pKa = 3.81LWGAMEE693 pKa = 5.26DD694 pKa = 3.55SGLTTEE700 pKa = 5.24LALNPEE706 pKa = 4.64FSMTHH711 pKa = 5.57EE712 pKa = 4.42LLTRR716 pKa = 11.84LNTPKK721 pKa = 9.08QTSPEE726 pKa = 3.94HH727 pKa = 6.1MPVKK731 pKa = 10.12PKK733 pKa = 9.86PRR735 pKa = 11.84HH736 pKa = 4.71LSKK739 pKa = 10.91PEE741 pKa = 3.77LMADD745 pKa = 3.5LQEE748 pKa = 4.51LKK750 pKa = 10.77KK751 pKa = 11.03SLTNLTSSHH760 pKa = 5.29TEE762 pKa = 3.48KK763 pKa = 11.08LLAKK767 pKa = 9.86PEE769 pKa = 4.09YY770 pKa = 10.07VRR772 pKa = 11.84SKK774 pKa = 10.76LEE776 pKa = 3.89EE777 pKa = 4.1VIKK780 pKa = 9.49TFPSLEE786 pKa = 4.26AIITIMGKK794 pKa = 9.98YY795 pKa = 9.07EE796 pKa = 4.03DD797 pKa = 5.18LYY799 pKa = 11.51SEE801 pKa = 4.75LDD803 pKa = 3.25EE804 pKa = 4.83VYY806 pKa = 10.58NNKK809 pKa = 9.0MKK811 pKa = 10.57IEE813 pKa = 4.28DD814 pKa = 3.86FMDD817 pKa = 5.25KK818 pKa = 9.45LTHH821 pKa = 7.26WINQAQHH828 pKa = 5.33EE829 pKa = 4.66MEE831 pKa = 3.84EE832 pKa = 4.25AMRR835 pKa = 11.84KK836 pKa = 9.33SIRR839 pKa = 11.84VVQQAQVQDD848 pKa = 3.93YY849 pKa = 9.93EE850 pKa = 4.52NIEE853 pKa = 3.88AFYY856 pKa = 10.93EE857 pKa = 4.1KK858 pKa = 10.65DD859 pKa = 3.24PTKK862 pKa = 10.7LADD865 pKa = 3.53PLGYY869 pKa = 10.26KK870 pKa = 10.15GAQTLVGKK878 pKa = 8.49HH879 pKa = 5.41TFPEE883 pKa = 4.43SSSTIPASLTKK894 pKa = 10.28SQKK897 pKa = 10.57KK898 pKa = 9.08NLKK901 pKa = 9.19KK902 pKa = 10.14KK903 pKa = 10.08QRR905 pKa = 11.84ISKK908 pKa = 10.14LRR910 pKa = 11.84DD911 pKa = 2.81IEE913 pKa = 4.14QASWSRR919 pKa = 11.84DD920 pKa = 3.16ALGEE924 pKa = 4.0IRR926 pKa = 11.84KK927 pKa = 9.41HH928 pKa = 6.43EE929 pKa = 4.23EE930 pKa = 3.74DD931 pKa = 3.5LL932 pKa = 4.52
Molecular weight: 107.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2064 |
153 |
979 |
688.0 |
77.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.444 ± 0.605 | 0.969 ± 0.242 |
5.378 ± 0.444 | 5.959 ± 0.818 |
2.665 ± 0.746 | 5.959 ± 1.208 |
1.89 ± 0.336 | 6.541 ± 0.774 |
6.153 ± 1.126 | 8.915 ± 0.998 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.616 ± 0.278 | 5.039 ± 0.589 |
6.105 ± 0.667 | 4.36 ± 0.532 |
5.087 ± 0.05 | 7.219 ± 1.132 |
7.897 ± 0.454 | 5.329 ± 0.986 |
0.969 ± 0.24 | 4.506 ± 0.103 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
