
Beihai weivirus-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.66
Get precalculated fractions of proteins
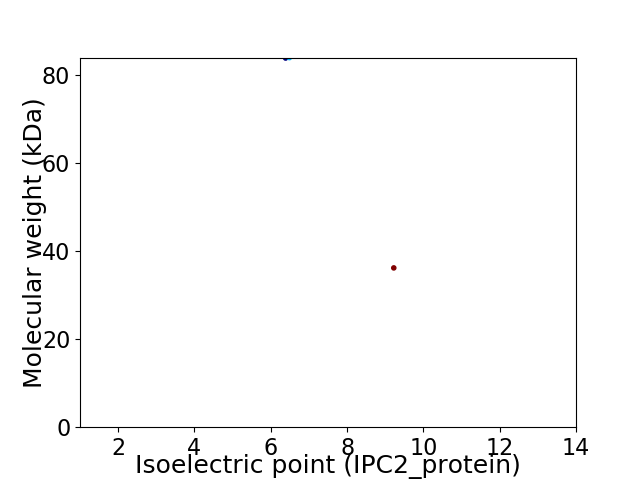
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL57|A0A1L3KL57_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 7 OX=1922754 PE=4 SV=1
MM1 pKa = 7.58LPFSLDD7 pKa = 2.77RR8 pKa = 11.84SYY10 pKa = 11.73RR11 pKa = 11.84EE12 pKa = 4.02LFLVEE17 pKa = 3.3PWIRR21 pKa = 11.84EE22 pKa = 3.9VASNRR27 pKa = 11.84EE28 pKa = 3.65GDD30 pKa = 3.71YY31 pKa = 11.05KK32 pKa = 10.46LTVFARR38 pKa = 11.84HH39 pKa = 5.36WFPWLWCSKK48 pKa = 9.55KK49 pKa = 10.54QNAEE53 pKa = 3.54AFSIVFTGKK62 pKa = 9.0EE63 pKa = 3.68WSSLSFCIEE72 pKa = 3.63HH73 pKa = 7.41ALRR76 pKa = 11.84TGLKK80 pKa = 9.17EE81 pKa = 3.82ANAYY85 pKa = 9.43GAFVSKK91 pKa = 10.42AQQLWPGGSQEE102 pKa = 3.97EE103 pKa = 4.72VAMKK107 pKa = 9.67MLAPHH112 pKa = 6.37MVLQYY117 pKa = 9.19LTYY120 pKa = 9.86KK121 pKa = 10.27RR122 pKa = 11.84VVVDD126 pKa = 3.51HH127 pKa = 7.07CCFSLRR133 pKa = 11.84YY134 pKa = 9.86SPFLKK139 pKa = 10.65PLDD142 pKa = 3.48RR143 pKa = 11.84TKK145 pKa = 11.31NKK147 pKa = 8.94TVEE150 pKa = 3.8ATKK153 pKa = 10.18PDD155 pKa = 3.64KK156 pKa = 10.84KK157 pKa = 10.63DD158 pKa = 3.43GQGKK162 pKa = 9.53SEE164 pKa = 4.82GGGTDD169 pKa = 4.46DD170 pKa = 4.03NPKK173 pKa = 10.64SGGEE177 pKa = 3.84KK178 pKa = 10.46SPVQPHH184 pKa = 5.81VPHH187 pKa = 6.65VDD189 pKa = 3.27AEE191 pKa = 4.23EE192 pKa = 3.98QPIVDD197 pKa = 4.46PAPGGTRR204 pKa = 11.84MADD207 pKa = 3.03TGSCYY212 pKa = 10.52AAIVGQEE219 pKa = 4.13DD220 pKa = 3.64GHH222 pKa = 7.36ARR224 pKa = 11.84TVDD227 pKa = 3.58GEE229 pKa = 4.02PDD231 pKa = 3.08IVGGVLLPISRR242 pKa = 11.84EE243 pKa = 3.97PNVPSDD249 pKa = 3.74TINNVKK255 pKa = 10.23KK256 pKa = 10.57AIKK259 pKa = 9.85EE260 pKa = 4.08RR261 pKa = 11.84IDD263 pKa = 3.96EE264 pKa = 4.32KK265 pKa = 11.07QLPCNLTKK273 pKa = 10.91DD274 pKa = 3.32EE275 pKa = 4.07TRR277 pKa = 11.84RR278 pKa = 11.84IGRR281 pKa = 11.84LVSASLADD289 pKa = 3.52NGPFARR295 pKa = 11.84KK296 pKa = 9.94RR297 pKa = 11.84IRR299 pKa = 11.84AWLAKK304 pKa = 10.34NFDD307 pKa = 3.75LAEE310 pKa = 4.34IKK312 pKa = 10.23SKK314 pKa = 10.14KK315 pKa = 8.58WSEE318 pKa = 3.79EE319 pKa = 3.71RR320 pKa = 11.84LRR322 pKa = 11.84AAVDD326 pKa = 3.43KK327 pKa = 10.73LYY329 pKa = 10.1STSDD333 pKa = 3.37PQFSYY338 pKa = 11.04AAAVKK343 pKa = 10.05AEE345 pKa = 4.0QMPEE349 pKa = 3.89GKK351 pKa = 9.34PPRR354 pKa = 11.84MLIADD359 pKa = 4.64GDD361 pKa = 4.27PGQVMALMTIACFEE375 pKa = 4.19SLMYY379 pKa = 10.08EE380 pKa = 3.99WYY382 pKa = 9.45EE383 pKa = 3.81NLSIKK388 pKa = 10.0HH389 pKa = 5.57ASRR392 pKa = 11.84RR393 pKa = 11.84AAMKK397 pKa = 10.15RR398 pKa = 11.84VLNHH402 pKa = 6.28LRR404 pKa = 11.84QDD406 pKa = 3.93SEE408 pKa = 4.75CSFLEE413 pKa = 4.31GDD415 pKa = 3.85GSAWDD420 pKa = 4.02TTCSHH425 pKa = 7.04RR426 pKa = 11.84VRR428 pKa = 11.84SIVEE432 pKa = 3.79NPVLKK437 pKa = 10.6RR438 pKa = 11.84ILEE441 pKa = 4.24VARR444 pKa = 11.84EE445 pKa = 3.91QAVVPEE451 pKa = 4.27QWLEE455 pKa = 3.84AHH457 pKa = 6.59DD458 pKa = 3.87TSTSKK463 pKa = 10.68KK464 pKa = 9.43HH465 pKa = 6.34LKK467 pKa = 10.49LKK469 pKa = 9.31LTEE472 pKa = 4.26KK473 pKa = 9.57GKK475 pKa = 10.69KK476 pKa = 8.1MVYY479 pKa = 9.81EE480 pKa = 4.5VIDD483 pKa = 3.81SIRR486 pKa = 11.84RR487 pKa = 11.84SGHH490 pKa = 6.33RR491 pKa = 11.84GTSCLNWWINNVLWICALVQNPEE514 pKa = 3.8KK515 pKa = 10.69SISATATNFMDD526 pKa = 4.54FWGEE530 pKa = 3.87TITIKK535 pKa = 10.58RR536 pKa = 11.84AFEE539 pKa = 4.49GDD541 pKa = 3.56DD542 pKa = 3.4SGLTVAPKK550 pKa = 10.07MKK552 pKa = 10.15DD553 pKa = 3.2VKK555 pKa = 10.89DD556 pKa = 3.55PRR558 pKa = 11.84FEE560 pKa = 4.04HH561 pKa = 7.03ALEE564 pKa = 4.24FWHH567 pKa = 7.23RR568 pKa = 11.84AGFNMKK574 pKa = 9.63IFLRR578 pKa = 11.84HH579 pKa = 4.9NVGLFVGTEE588 pKa = 3.64IALDD592 pKa = 3.46EE593 pKa = 4.89HH594 pKa = 6.94GPTGEE599 pKa = 4.12YY600 pKa = 10.63APEE603 pKa = 4.12LKK605 pKa = 9.57RR606 pKa = 11.84TFEE609 pKa = 4.21KK610 pKa = 10.96AGISCSSLTKK620 pKa = 10.64KK621 pKa = 10.45LIKK624 pKa = 10.53AGAAGNEE631 pKa = 4.13GLHH634 pKa = 4.98EE635 pKa = 4.29VRR637 pKa = 11.84RR638 pKa = 11.84SLALAKK644 pKa = 10.36AYY646 pKa = 10.29DD647 pKa = 3.72FAGIVPSVSRR657 pKa = 11.84KK658 pKa = 9.66YY659 pKa = 10.54LLCAQGLDD667 pKa = 3.98TMDD670 pKa = 5.08HH671 pKa = 5.48EE672 pKa = 5.05LKK674 pKa = 10.09MRR676 pKa = 11.84TGEE679 pKa = 4.16TTLEE683 pKa = 4.7GIRR686 pKa = 11.84DD687 pKa = 4.03MIDD690 pKa = 3.12AANSTCSPEE699 pKa = 5.46DD700 pKa = 3.4EE701 pKa = 4.79DD702 pKa = 5.81ALLEE706 pKa = 4.0RR707 pKa = 11.84LGRR710 pKa = 11.84SISQTEE716 pKa = 3.53RR717 pKa = 11.84DD718 pKa = 3.79AFEE721 pKa = 4.41AYY723 pKa = 9.0CWAWEE728 pKa = 4.16KK729 pKa = 10.74EE730 pKa = 4.09ATSQCEE736 pKa = 3.84EE737 pKa = 4.21FAASLPASWRR747 pKa = 11.84VV748 pKa = 3.01
MM1 pKa = 7.58LPFSLDD7 pKa = 2.77RR8 pKa = 11.84SYY10 pKa = 11.73RR11 pKa = 11.84EE12 pKa = 4.02LFLVEE17 pKa = 3.3PWIRR21 pKa = 11.84EE22 pKa = 3.9VASNRR27 pKa = 11.84EE28 pKa = 3.65GDD30 pKa = 3.71YY31 pKa = 11.05KK32 pKa = 10.46LTVFARR38 pKa = 11.84HH39 pKa = 5.36WFPWLWCSKK48 pKa = 9.55KK49 pKa = 10.54QNAEE53 pKa = 3.54AFSIVFTGKK62 pKa = 9.0EE63 pKa = 3.68WSSLSFCIEE72 pKa = 3.63HH73 pKa = 7.41ALRR76 pKa = 11.84TGLKK80 pKa = 9.17EE81 pKa = 3.82ANAYY85 pKa = 9.43GAFVSKK91 pKa = 10.42AQQLWPGGSQEE102 pKa = 3.97EE103 pKa = 4.72VAMKK107 pKa = 9.67MLAPHH112 pKa = 6.37MVLQYY117 pKa = 9.19LTYY120 pKa = 9.86KK121 pKa = 10.27RR122 pKa = 11.84VVVDD126 pKa = 3.51HH127 pKa = 7.07CCFSLRR133 pKa = 11.84YY134 pKa = 9.86SPFLKK139 pKa = 10.65PLDD142 pKa = 3.48RR143 pKa = 11.84TKK145 pKa = 11.31NKK147 pKa = 8.94TVEE150 pKa = 3.8ATKK153 pKa = 10.18PDD155 pKa = 3.64KK156 pKa = 10.84KK157 pKa = 10.63DD158 pKa = 3.43GQGKK162 pKa = 9.53SEE164 pKa = 4.82GGGTDD169 pKa = 4.46DD170 pKa = 4.03NPKK173 pKa = 10.64SGGEE177 pKa = 3.84KK178 pKa = 10.46SPVQPHH184 pKa = 5.81VPHH187 pKa = 6.65VDD189 pKa = 3.27AEE191 pKa = 4.23EE192 pKa = 3.98QPIVDD197 pKa = 4.46PAPGGTRR204 pKa = 11.84MADD207 pKa = 3.03TGSCYY212 pKa = 10.52AAIVGQEE219 pKa = 4.13DD220 pKa = 3.64GHH222 pKa = 7.36ARR224 pKa = 11.84TVDD227 pKa = 3.58GEE229 pKa = 4.02PDD231 pKa = 3.08IVGGVLLPISRR242 pKa = 11.84EE243 pKa = 3.97PNVPSDD249 pKa = 3.74TINNVKK255 pKa = 10.23KK256 pKa = 10.57AIKK259 pKa = 9.85EE260 pKa = 4.08RR261 pKa = 11.84IDD263 pKa = 3.96EE264 pKa = 4.32KK265 pKa = 11.07QLPCNLTKK273 pKa = 10.91DD274 pKa = 3.32EE275 pKa = 4.07TRR277 pKa = 11.84RR278 pKa = 11.84IGRR281 pKa = 11.84LVSASLADD289 pKa = 3.52NGPFARR295 pKa = 11.84KK296 pKa = 9.94RR297 pKa = 11.84IRR299 pKa = 11.84AWLAKK304 pKa = 10.34NFDD307 pKa = 3.75LAEE310 pKa = 4.34IKK312 pKa = 10.23SKK314 pKa = 10.14KK315 pKa = 8.58WSEE318 pKa = 3.79EE319 pKa = 3.71RR320 pKa = 11.84LRR322 pKa = 11.84AAVDD326 pKa = 3.43KK327 pKa = 10.73LYY329 pKa = 10.1STSDD333 pKa = 3.37PQFSYY338 pKa = 11.04AAAVKK343 pKa = 10.05AEE345 pKa = 4.0QMPEE349 pKa = 3.89GKK351 pKa = 9.34PPRR354 pKa = 11.84MLIADD359 pKa = 4.64GDD361 pKa = 4.27PGQVMALMTIACFEE375 pKa = 4.19SLMYY379 pKa = 10.08EE380 pKa = 3.99WYY382 pKa = 9.45EE383 pKa = 3.81NLSIKK388 pKa = 10.0HH389 pKa = 5.57ASRR392 pKa = 11.84RR393 pKa = 11.84AAMKK397 pKa = 10.15RR398 pKa = 11.84VLNHH402 pKa = 6.28LRR404 pKa = 11.84QDD406 pKa = 3.93SEE408 pKa = 4.75CSFLEE413 pKa = 4.31GDD415 pKa = 3.85GSAWDD420 pKa = 4.02TTCSHH425 pKa = 7.04RR426 pKa = 11.84VRR428 pKa = 11.84SIVEE432 pKa = 3.79NPVLKK437 pKa = 10.6RR438 pKa = 11.84ILEE441 pKa = 4.24VARR444 pKa = 11.84EE445 pKa = 3.91QAVVPEE451 pKa = 4.27QWLEE455 pKa = 3.84AHH457 pKa = 6.59DD458 pKa = 3.87TSTSKK463 pKa = 10.68KK464 pKa = 9.43HH465 pKa = 6.34LKK467 pKa = 10.49LKK469 pKa = 9.31LTEE472 pKa = 4.26KK473 pKa = 9.57GKK475 pKa = 10.69KK476 pKa = 8.1MVYY479 pKa = 9.81EE480 pKa = 4.5VIDD483 pKa = 3.81SIRR486 pKa = 11.84RR487 pKa = 11.84SGHH490 pKa = 6.33RR491 pKa = 11.84GTSCLNWWINNVLWICALVQNPEE514 pKa = 3.8KK515 pKa = 10.69SISATATNFMDD526 pKa = 4.54FWGEE530 pKa = 3.87TITIKK535 pKa = 10.58RR536 pKa = 11.84AFEE539 pKa = 4.49GDD541 pKa = 3.56DD542 pKa = 3.4SGLTVAPKK550 pKa = 10.07MKK552 pKa = 10.15DD553 pKa = 3.2VKK555 pKa = 10.89DD556 pKa = 3.55PRR558 pKa = 11.84FEE560 pKa = 4.04HH561 pKa = 7.03ALEE564 pKa = 4.24FWHH567 pKa = 7.23RR568 pKa = 11.84AGFNMKK574 pKa = 9.63IFLRR578 pKa = 11.84HH579 pKa = 4.9NVGLFVGTEE588 pKa = 3.64IALDD592 pKa = 3.46EE593 pKa = 4.89HH594 pKa = 6.94GPTGEE599 pKa = 4.12YY600 pKa = 10.63APEE603 pKa = 4.12LKK605 pKa = 9.57RR606 pKa = 11.84TFEE609 pKa = 4.21KK610 pKa = 10.96AGISCSSLTKK620 pKa = 10.64KK621 pKa = 10.45LIKK624 pKa = 10.53AGAAGNEE631 pKa = 4.13GLHH634 pKa = 4.98EE635 pKa = 4.29VRR637 pKa = 11.84RR638 pKa = 11.84SLALAKK644 pKa = 10.36AYY646 pKa = 10.29DD647 pKa = 3.72FAGIVPSVSRR657 pKa = 11.84KK658 pKa = 9.66YY659 pKa = 10.54LLCAQGLDD667 pKa = 3.98TMDD670 pKa = 5.08HH671 pKa = 5.48EE672 pKa = 5.05LKK674 pKa = 10.09MRR676 pKa = 11.84TGEE679 pKa = 4.16TTLEE683 pKa = 4.7GIRR686 pKa = 11.84DD687 pKa = 4.03MIDD690 pKa = 3.12AANSTCSPEE699 pKa = 5.46DD700 pKa = 3.4EE701 pKa = 4.79DD702 pKa = 5.81ALLEE706 pKa = 4.0RR707 pKa = 11.84LGRR710 pKa = 11.84SISQTEE716 pKa = 3.53RR717 pKa = 11.84DD718 pKa = 3.79AFEE721 pKa = 4.41AYY723 pKa = 9.0CWAWEE728 pKa = 4.16KK729 pKa = 10.74EE730 pKa = 4.09ATSQCEE736 pKa = 3.84EE737 pKa = 4.21FAASLPASWRR747 pKa = 11.84VV748 pKa = 3.01
Molecular weight: 83.97 kDa
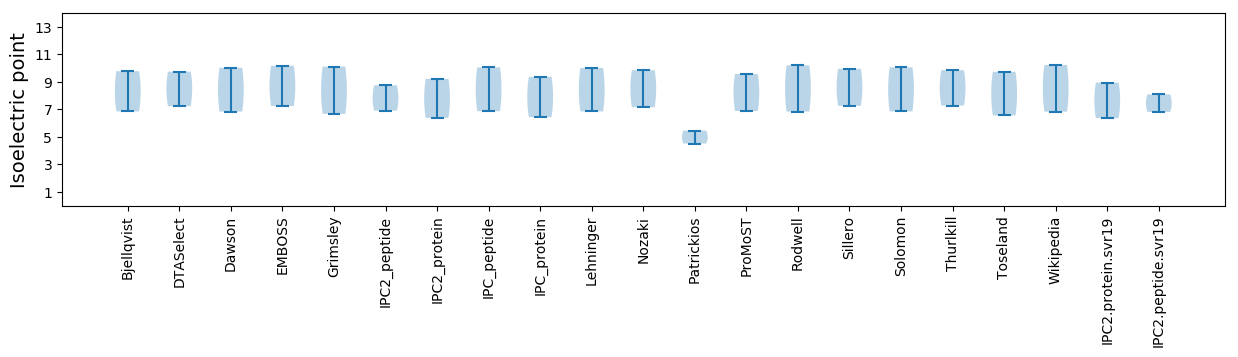
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL57|A0A1L3KL57_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 7 OX=1922754 PE=4 SV=1
MM1 pKa = 7.65ARR3 pKa = 11.84ARR5 pKa = 11.84KK6 pKa = 9.9NGTQARR12 pKa = 11.84KK13 pKa = 9.64RR14 pKa = 11.84MPRR17 pKa = 11.84RR18 pKa = 11.84GRR20 pKa = 11.84RR21 pKa = 11.84SGAPAVQAQGTGTTVAVPFGSNGRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 10.5SMAAMAMQGWNAFHH62 pKa = 7.57PYY64 pKa = 9.88HH65 pKa = 6.77LPLPRR70 pKa = 11.84AVGPYY75 pKa = 8.91TVIRR79 pKa = 11.84TSTLITNSHH88 pKa = 6.49KK89 pKa = 10.5FMQFGTFVDD98 pKa = 5.71DD99 pKa = 3.79NGFWTNVVGLGSVVSSDD116 pKa = 4.13PINGGGNTALFTVPTPAVASLSGTGFTCVPASLSVQVMNNTALMQANGIFGGGVCHH172 pKa = 6.05TQMSLGGRR180 pKa = 11.84AEE182 pKa = 4.36TYY184 pKa = 10.9NDD186 pKa = 3.71LSTEE190 pKa = 4.81FISYY194 pKa = 7.47MRR196 pKa = 11.84PRR198 pKa = 11.84LMSGGKK204 pKa = 9.21LALKK208 pKa = 9.49GVQMDD213 pKa = 4.59SYY215 pKa = 9.61PMNMSALANFEE226 pKa = 5.07KK227 pKa = 9.59ITKK230 pKa = 10.35NIDD233 pKa = 3.11HH234 pKa = 7.42NITYY238 pKa = 10.41SAGPYY243 pKa = 7.54ATGMAPIIFVNQSEE257 pKa = 4.61AEE259 pKa = 3.97ITYY262 pKa = 8.35LACIEE267 pKa = 3.87WRR269 pKa = 11.84VRR271 pKa = 11.84FDD273 pKa = 2.86IGSAAVASHH282 pKa = 5.33VHH284 pKa = 6.23HH285 pKa = 6.71GVTPDD290 pKa = 3.89SLWNDD295 pKa = 4.03CVQTAVSLGHH305 pKa = 5.75GVKK308 pKa = 10.39EE309 pKa = 4.18IADD312 pKa = 3.91TVATVGTSINNVRR325 pKa = 11.84AALFRR330 pKa = 11.84ANPQPMLVDD339 pKa = 3.8
MM1 pKa = 7.65ARR3 pKa = 11.84ARR5 pKa = 11.84KK6 pKa = 9.9NGTQARR12 pKa = 11.84KK13 pKa = 9.64RR14 pKa = 11.84MPRR17 pKa = 11.84RR18 pKa = 11.84GRR20 pKa = 11.84RR21 pKa = 11.84SGAPAVQAQGTGTTVAVPFGSNGRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 10.5SMAAMAMQGWNAFHH62 pKa = 7.57PYY64 pKa = 9.88HH65 pKa = 6.77LPLPRR70 pKa = 11.84AVGPYY75 pKa = 8.91TVIRR79 pKa = 11.84TSTLITNSHH88 pKa = 6.49KK89 pKa = 10.5FMQFGTFVDD98 pKa = 5.71DD99 pKa = 3.79NGFWTNVVGLGSVVSSDD116 pKa = 4.13PINGGGNTALFTVPTPAVASLSGTGFTCVPASLSVQVMNNTALMQANGIFGGGVCHH172 pKa = 6.05TQMSLGGRR180 pKa = 11.84AEE182 pKa = 4.36TYY184 pKa = 10.9NDD186 pKa = 3.71LSTEE190 pKa = 4.81FISYY194 pKa = 7.47MRR196 pKa = 11.84PRR198 pKa = 11.84LMSGGKK204 pKa = 9.21LALKK208 pKa = 9.49GVQMDD213 pKa = 4.59SYY215 pKa = 9.61PMNMSALANFEE226 pKa = 5.07KK227 pKa = 9.59ITKK230 pKa = 10.35NIDD233 pKa = 3.11HH234 pKa = 7.42NITYY238 pKa = 10.41SAGPYY243 pKa = 7.54ATGMAPIIFVNQSEE257 pKa = 4.61AEE259 pKa = 3.97ITYY262 pKa = 8.35LACIEE267 pKa = 3.87WRR269 pKa = 11.84VRR271 pKa = 11.84FDD273 pKa = 2.86IGSAAVASHH282 pKa = 5.33VHH284 pKa = 6.23HH285 pKa = 6.71GVTPDD290 pKa = 3.89SLWNDD295 pKa = 4.03CVQTAVSLGHH305 pKa = 5.75GVKK308 pKa = 10.39EE309 pKa = 4.18IADD312 pKa = 3.91TVATVGTSINNVRR325 pKa = 11.84AALFRR330 pKa = 11.84ANPQPMLVDD339 pKa = 3.8
Molecular weight: 36.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1087 |
339 |
748 |
543.5 |
60.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.66 ± 0.474 | 1.84 ± 0.326 |
4.876 ± 0.805 | 6.348 ± 2.115 |
3.772 ± 0.177 | 7.544 ± 1.227 |
2.576 ± 0.039 | 4.416 ± 0.004 |
5.888 ± 1.596 | 7.452 ± 0.912 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.128 ± 0.786 | 4.048 ± 1.06 |
4.968 ± 0.169 | 2.944 ± 0.294 |
6.164 ± 0.13 | 7.268 ± 0.053 |
6.072 ± 1.08 | 6.716 ± 0.908 |
2.116 ± 0.462 | 2.208 ± 0.075 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
