
Hubei sobemo-like virus 27
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
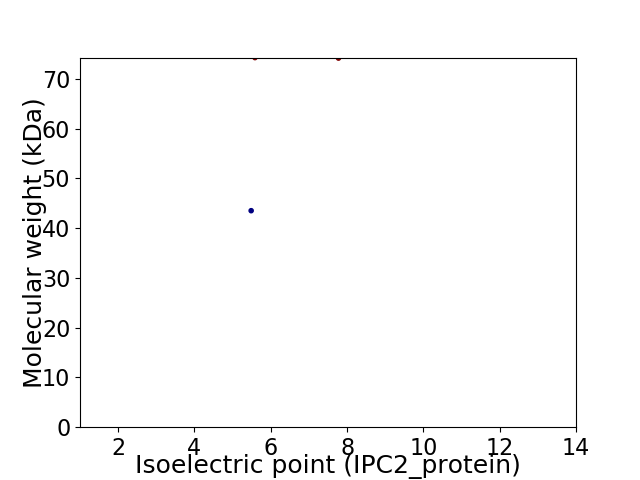
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
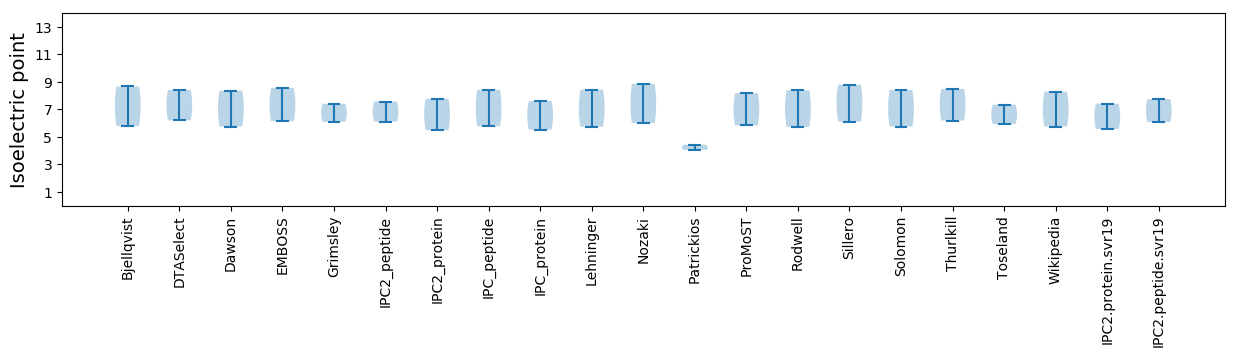
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KF61|A0A1L3KF61_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 27 OX=1923213 PE=4 SV=1
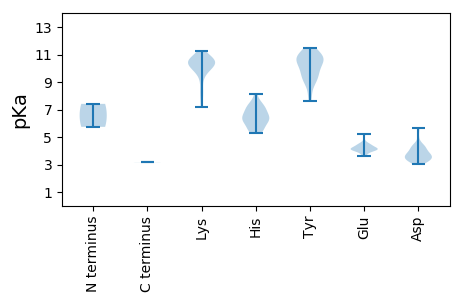
MM1 pKa = 5.74QTAVAKK7 pKa = 10.69HH8 pKa = 5.28EE9 pKa = 4.12AARR12 pKa = 11.84WDD14 pKa = 3.58IPDD17 pKa = 4.34DD18 pKa = 3.92FLEE21 pKa = 4.48EE22 pKa = 4.1THH24 pKa = 6.73FLRR27 pKa = 11.84AVEE30 pKa = 3.96EE31 pKa = 4.05VDD33 pKa = 3.77YY34 pKa = 10.72KK35 pKa = 11.22SSPGYY40 pKa = 9.81PYY42 pKa = 10.02MVRR45 pKa = 11.84NPSNSDD51 pKa = 3.09FFGVVDD57 pKa = 5.07GVANPARR64 pKa = 11.84LKK66 pKa = 10.01EE67 pKa = 3.65IWEE70 pKa = 4.03IVQIRR75 pKa = 11.84LHH77 pKa = 6.44KK78 pKa = 10.59LLLGEE83 pKa = 5.18DD84 pKa = 3.51DD85 pKa = 4.24CDD87 pKa = 4.42YY88 pKa = 11.35IRR90 pKa = 11.84LFIKK94 pKa = 10.68AEE96 pKa = 4.37PIKK99 pKa = 10.3EE100 pKa = 4.06KK101 pKa = 10.6KK102 pKa = 10.45LRR104 pKa = 11.84DD105 pKa = 3.35HH106 pKa = 7.54KK107 pKa = 11.15YY108 pKa = 9.69RR109 pKa = 11.84LISSVSVIDD118 pKa = 4.29QIIDD122 pKa = 3.27HH123 pKa = 6.32MLFDD127 pKa = 4.58PMNHH131 pKa = 6.29RR132 pKa = 11.84LYY134 pKa = 10.87DD135 pKa = 3.45NWHH138 pKa = 5.6MVPSKK143 pKa = 10.96VGWSPYY149 pKa = 9.14KK150 pKa = 10.42GGWRR154 pKa = 11.84IFPTQPLLAIDD165 pKa = 3.82KK166 pKa = 10.62SSWDD170 pKa = 3.21WTVCGWLIEE179 pKa = 4.27QIVQFRR185 pKa = 11.84IMLCNNINDD194 pKa = 3.72SWIKK198 pKa = 10.57LATQRR203 pKa = 11.84YY204 pKa = 8.12KK205 pKa = 11.02CLFLNPLFLTSGGLVLKK222 pKa = 10.48QKK224 pKa = 10.16EE225 pKa = 4.19AGVMKK230 pKa = 10.08SGCVNTIADD239 pKa = 3.89NSIMQLLLHH248 pKa = 6.37LRR250 pKa = 11.84VSLEE254 pKa = 3.72RR255 pKa = 11.84GGDD258 pKa = 3.48VDD260 pKa = 4.12GLFYY264 pKa = 10.89TMGDD268 pKa = 3.28DD269 pKa = 4.12TVQDD273 pKa = 4.14EE274 pKa = 4.71PNDD277 pKa = 3.72LDD279 pKa = 5.68DD280 pKa = 4.35YY281 pKa = 11.48LSRR284 pKa = 11.84LNQFSIVKK292 pKa = 9.54QATNRR297 pKa = 11.84TEE299 pKa = 3.77FAGMHH304 pKa = 6.09FNYY307 pKa = 10.44SQVEE311 pKa = 3.7PMYY314 pKa = 10.62GGKK317 pKa = 9.13HH318 pKa = 5.48AFNLLHH324 pKa = 7.18LDD326 pKa = 3.86PEE328 pKa = 4.45NEE330 pKa = 4.06EE331 pKa = 4.47QVCSSYY337 pKa = 10.73MLLYY341 pKa = 10.32HH342 pKa = 6.92RR343 pKa = 11.84SKK345 pKa = 10.63FRR347 pKa = 11.84GWFEE351 pKa = 4.62SLFEE355 pKa = 4.32AMNIPVPHH363 pKa = 6.67RR364 pKa = 11.84LVRR367 pKa = 11.84DD368 pKa = 4.0HH369 pKa = 7.28IFDD372 pKa = 3.55NSS374 pKa = 3.16
MM1 pKa = 5.74QTAVAKK7 pKa = 10.69HH8 pKa = 5.28EE9 pKa = 4.12AARR12 pKa = 11.84WDD14 pKa = 3.58IPDD17 pKa = 4.34DD18 pKa = 3.92FLEE21 pKa = 4.48EE22 pKa = 4.1THH24 pKa = 6.73FLRR27 pKa = 11.84AVEE30 pKa = 3.96EE31 pKa = 4.05VDD33 pKa = 3.77YY34 pKa = 10.72KK35 pKa = 11.22SSPGYY40 pKa = 9.81PYY42 pKa = 10.02MVRR45 pKa = 11.84NPSNSDD51 pKa = 3.09FFGVVDD57 pKa = 5.07GVANPARR64 pKa = 11.84LKK66 pKa = 10.01EE67 pKa = 3.65IWEE70 pKa = 4.03IVQIRR75 pKa = 11.84LHH77 pKa = 6.44KK78 pKa = 10.59LLLGEE83 pKa = 5.18DD84 pKa = 3.51DD85 pKa = 4.24CDD87 pKa = 4.42YY88 pKa = 11.35IRR90 pKa = 11.84LFIKK94 pKa = 10.68AEE96 pKa = 4.37PIKK99 pKa = 10.3EE100 pKa = 4.06KK101 pKa = 10.6KK102 pKa = 10.45LRR104 pKa = 11.84DD105 pKa = 3.35HH106 pKa = 7.54KK107 pKa = 11.15YY108 pKa = 9.69RR109 pKa = 11.84LISSVSVIDD118 pKa = 4.29QIIDD122 pKa = 3.27HH123 pKa = 6.32MLFDD127 pKa = 4.58PMNHH131 pKa = 6.29RR132 pKa = 11.84LYY134 pKa = 10.87DD135 pKa = 3.45NWHH138 pKa = 5.6MVPSKK143 pKa = 10.96VGWSPYY149 pKa = 9.14KK150 pKa = 10.42GGWRR154 pKa = 11.84IFPTQPLLAIDD165 pKa = 3.82KK166 pKa = 10.62SSWDD170 pKa = 3.21WTVCGWLIEE179 pKa = 4.27QIVQFRR185 pKa = 11.84IMLCNNINDD194 pKa = 3.72SWIKK198 pKa = 10.57LATQRR203 pKa = 11.84YY204 pKa = 8.12KK205 pKa = 11.02CLFLNPLFLTSGGLVLKK222 pKa = 10.48QKK224 pKa = 10.16EE225 pKa = 4.19AGVMKK230 pKa = 10.08SGCVNTIADD239 pKa = 3.89NSIMQLLLHH248 pKa = 6.37LRR250 pKa = 11.84VSLEE254 pKa = 3.72RR255 pKa = 11.84GGDD258 pKa = 3.48VDD260 pKa = 4.12GLFYY264 pKa = 10.89TMGDD268 pKa = 3.28DD269 pKa = 4.12TVQDD273 pKa = 4.14EE274 pKa = 4.71PNDD277 pKa = 3.72LDD279 pKa = 5.68DD280 pKa = 4.35YY281 pKa = 11.48LSRR284 pKa = 11.84LNQFSIVKK292 pKa = 9.54QATNRR297 pKa = 11.84TEE299 pKa = 3.77FAGMHH304 pKa = 6.09FNYY307 pKa = 10.44SQVEE311 pKa = 3.7PMYY314 pKa = 10.62GGKK317 pKa = 9.13HH318 pKa = 5.48AFNLLHH324 pKa = 7.18LDD326 pKa = 3.86PEE328 pKa = 4.45NEE330 pKa = 4.06EE331 pKa = 4.47QVCSSYY337 pKa = 10.73MLLYY341 pKa = 10.32HH342 pKa = 6.92RR343 pKa = 11.84SKK345 pKa = 10.63FRR347 pKa = 11.84GWFEE351 pKa = 4.62SLFEE355 pKa = 4.32AMNIPVPHH363 pKa = 6.67RR364 pKa = 11.84LVRR367 pKa = 11.84DD368 pKa = 4.0HH369 pKa = 7.28IFDD372 pKa = 3.55NSS374 pKa = 3.16
Molecular weight: 43.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF61|A0A1L3KF61_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 27 OX=1923213 PE=4 SV=1
MM1 pKa = 7.41NPIILEE7 pKa = 4.07NEE9 pKa = 3.97EE10 pKa = 4.02TQEE13 pKa = 3.94NVLLEE18 pKa = 4.14RR19 pKa = 11.84TPQRR23 pKa = 11.84SFINRR28 pKa = 11.84IFWKK32 pKa = 9.92VVVVFATLLSLTALIPLLWLVIRR55 pKa = 11.84ALTYY59 pKa = 9.03RR60 pKa = 11.84LYY62 pKa = 11.12VAVEE66 pKa = 4.11YY67 pKa = 10.7QVVTQYY73 pKa = 10.51EE74 pKa = 4.24ALWRR78 pKa = 11.84FLAGLGQDD86 pKa = 4.23PPPEE90 pKa = 4.11PSVIEE95 pKa = 4.08KK96 pKa = 10.63VADD99 pKa = 3.44HH100 pKa = 5.73VTDD103 pKa = 4.26MIRR106 pKa = 11.84RR107 pKa = 11.84IDD109 pKa = 3.72SNIIAIGVIGIITFLAVVVSTFFVLRR135 pKa = 11.84SLRR138 pKa = 11.84KK139 pKa = 7.52ISYY142 pKa = 9.52RR143 pKa = 11.84IRR145 pKa = 11.84GIQFEE150 pKa = 4.11ALRR153 pKa = 11.84PGSTYY158 pKa = 10.06TKK160 pKa = 10.61EE161 pKa = 4.19EE162 pKa = 3.75IPNFQVRR169 pKa = 11.84VGQPGLFSASHH180 pKa = 5.28VGYY183 pKa = 10.47AVRR186 pKa = 11.84IGPNYY191 pKa = 8.69MAVPHH196 pKa = 5.8HH197 pKa = 6.62VIRR200 pKa = 11.84GQSTVLLTGPKK211 pKa = 9.54GRR213 pKa = 11.84CIVDD217 pKa = 3.4VVATQSRR224 pKa = 11.84MYY226 pKa = 10.59TDD228 pKa = 5.63LSYY231 pKa = 11.17CWVDD235 pKa = 3.22EE236 pKa = 4.56RR237 pKa = 11.84VWTTLGISIAKK248 pKa = 7.8LQPYY252 pKa = 7.64TSRR255 pKa = 11.84TVVRR259 pKa = 11.84CYY261 pKa = 10.84GQLGYY266 pKa = 9.48TSGSIYY272 pKa = 9.33KK273 pKa = 9.6TSLSPGIIAYY283 pKa = 8.99EE284 pKa = 4.18GSTEE288 pKa = 4.02NGTSGAAYY296 pKa = 9.91VLAGKK301 pKa = 10.17VLGLHH306 pKa = 6.15SAHH309 pKa = 6.31NPSSNMNFGVSSNMLLTEE327 pKa = 4.16YY328 pKa = 11.26AMLMKK333 pKa = 10.53PEE335 pKa = 4.21GKK337 pKa = 8.3KK338 pKa = 9.0TSATLDD344 pKa = 3.8HH345 pKa = 6.39YY346 pKa = 10.14TGDD349 pKa = 3.6APVTNVGKK357 pKa = 10.02GWSISEE363 pKa = 4.07SMAKK367 pKa = 9.9KK368 pKa = 8.23EE369 pKa = 4.52QEE371 pKa = 3.98WNDD374 pKa = 3.41VKK376 pKa = 11.15LKK378 pKa = 10.78NLATAAYY385 pKa = 9.32SHH387 pKa = 7.48DD388 pKa = 3.4WTNPPEE394 pKa = 4.3IDD396 pKa = 3.66YY397 pKa = 10.81NANLDD402 pKa = 3.55WGSDD406 pKa = 3.71DD407 pKa = 4.43EE408 pKa = 4.93EE409 pKa = 4.39SAKK412 pKa = 10.54VKK414 pKa = 10.67RR415 pKa = 11.84PVFQAGAFVPATVTLQHH432 pKa = 6.05QGAEE436 pKa = 4.16GEE438 pKa = 4.34MVEE441 pKa = 4.26TGLMNVNLVEE451 pKa = 4.21MVGALSKK458 pKa = 10.56RR459 pKa = 11.84VEE461 pKa = 4.21EE462 pKa = 4.45LEE464 pKa = 4.34KK465 pKa = 10.52FCASITEE472 pKa = 4.16PVQRR476 pKa = 11.84LIEE479 pKa = 4.51DD480 pKa = 3.22VDD482 pKa = 3.63GLKK485 pKa = 10.3NRR487 pKa = 11.84KK488 pKa = 8.45VVNATAKK495 pKa = 10.34AVAKK499 pKa = 7.17VTCEE503 pKa = 3.98CGVEE507 pKa = 4.16VAEE510 pKa = 5.19GKK512 pKa = 10.46LEE514 pKa = 3.84NHH516 pKa = 7.05ILNAHH521 pKa = 7.08PKK523 pKa = 9.77PIVQMACPDD532 pKa = 3.72CHH534 pKa = 8.13VMVAAFKK541 pKa = 10.14MDD543 pKa = 3.2AHH545 pKa = 7.23IKK547 pKa = 9.85NSHH550 pKa = 6.16PPSTKK555 pKa = 10.02VKK557 pKa = 10.55CSFCEE562 pKa = 4.13IQCKK566 pKa = 10.29DD567 pKa = 3.35SNRR570 pKa = 11.84LANHH574 pKa = 6.75IANVHH579 pKa = 5.46RR580 pKa = 11.84VMEE583 pKa = 4.42VKK585 pKa = 10.42PEE587 pKa = 4.04SAFSSDD593 pKa = 3.05EE594 pKa = 4.3TIKK597 pKa = 10.97VATRR601 pKa = 11.84PFLGRR606 pKa = 11.84SQRR609 pKa = 11.84PRR611 pKa = 11.84SNSPRR616 pKa = 11.84MRR618 pKa = 11.84PQQLSRR624 pKa = 11.84TSNVSTKK631 pKa = 10.03SQNSPSLEE639 pKa = 3.95AVLSLMTASLQNIEE653 pKa = 4.45KK654 pKa = 10.24RR655 pKa = 11.84LNAKK659 pKa = 10.01PPGTDD664 pKa = 3.35GQSSGIGPNN673 pKa = 3.15
MM1 pKa = 7.41NPIILEE7 pKa = 4.07NEE9 pKa = 3.97EE10 pKa = 4.02TQEE13 pKa = 3.94NVLLEE18 pKa = 4.14RR19 pKa = 11.84TPQRR23 pKa = 11.84SFINRR28 pKa = 11.84IFWKK32 pKa = 9.92VVVVFATLLSLTALIPLLWLVIRR55 pKa = 11.84ALTYY59 pKa = 9.03RR60 pKa = 11.84LYY62 pKa = 11.12VAVEE66 pKa = 4.11YY67 pKa = 10.7QVVTQYY73 pKa = 10.51EE74 pKa = 4.24ALWRR78 pKa = 11.84FLAGLGQDD86 pKa = 4.23PPPEE90 pKa = 4.11PSVIEE95 pKa = 4.08KK96 pKa = 10.63VADD99 pKa = 3.44HH100 pKa = 5.73VTDD103 pKa = 4.26MIRR106 pKa = 11.84RR107 pKa = 11.84IDD109 pKa = 3.72SNIIAIGVIGIITFLAVVVSTFFVLRR135 pKa = 11.84SLRR138 pKa = 11.84KK139 pKa = 7.52ISYY142 pKa = 9.52RR143 pKa = 11.84IRR145 pKa = 11.84GIQFEE150 pKa = 4.11ALRR153 pKa = 11.84PGSTYY158 pKa = 10.06TKK160 pKa = 10.61EE161 pKa = 4.19EE162 pKa = 3.75IPNFQVRR169 pKa = 11.84VGQPGLFSASHH180 pKa = 5.28VGYY183 pKa = 10.47AVRR186 pKa = 11.84IGPNYY191 pKa = 8.69MAVPHH196 pKa = 5.8HH197 pKa = 6.62VIRR200 pKa = 11.84GQSTVLLTGPKK211 pKa = 9.54GRR213 pKa = 11.84CIVDD217 pKa = 3.4VVATQSRR224 pKa = 11.84MYY226 pKa = 10.59TDD228 pKa = 5.63LSYY231 pKa = 11.17CWVDD235 pKa = 3.22EE236 pKa = 4.56RR237 pKa = 11.84VWTTLGISIAKK248 pKa = 7.8LQPYY252 pKa = 7.64TSRR255 pKa = 11.84TVVRR259 pKa = 11.84CYY261 pKa = 10.84GQLGYY266 pKa = 9.48TSGSIYY272 pKa = 9.33KK273 pKa = 9.6TSLSPGIIAYY283 pKa = 8.99EE284 pKa = 4.18GSTEE288 pKa = 4.02NGTSGAAYY296 pKa = 9.91VLAGKK301 pKa = 10.17VLGLHH306 pKa = 6.15SAHH309 pKa = 6.31NPSSNMNFGVSSNMLLTEE327 pKa = 4.16YY328 pKa = 11.26AMLMKK333 pKa = 10.53PEE335 pKa = 4.21GKK337 pKa = 8.3KK338 pKa = 9.0TSATLDD344 pKa = 3.8HH345 pKa = 6.39YY346 pKa = 10.14TGDD349 pKa = 3.6APVTNVGKK357 pKa = 10.02GWSISEE363 pKa = 4.07SMAKK367 pKa = 9.9KK368 pKa = 8.23EE369 pKa = 4.52QEE371 pKa = 3.98WNDD374 pKa = 3.41VKK376 pKa = 11.15LKK378 pKa = 10.78NLATAAYY385 pKa = 9.32SHH387 pKa = 7.48DD388 pKa = 3.4WTNPPEE394 pKa = 4.3IDD396 pKa = 3.66YY397 pKa = 10.81NANLDD402 pKa = 3.55WGSDD406 pKa = 3.71DD407 pKa = 4.43EE408 pKa = 4.93EE409 pKa = 4.39SAKK412 pKa = 10.54VKK414 pKa = 10.67RR415 pKa = 11.84PVFQAGAFVPATVTLQHH432 pKa = 6.05QGAEE436 pKa = 4.16GEE438 pKa = 4.34MVEE441 pKa = 4.26TGLMNVNLVEE451 pKa = 4.21MVGALSKK458 pKa = 10.56RR459 pKa = 11.84VEE461 pKa = 4.21EE462 pKa = 4.45LEE464 pKa = 4.34KK465 pKa = 10.52FCASITEE472 pKa = 4.16PVQRR476 pKa = 11.84LIEE479 pKa = 4.51DD480 pKa = 3.22VDD482 pKa = 3.63GLKK485 pKa = 10.3NRR487 pKa = 11.84KK488 pKa = 8.45VVNATAKK495 pKa = 10.34AVAKK499 pKa = 7.17VTCEE503 pKa = 3.98CGVEE507 pKa = 4.16VAEE510 pKa = 5.19GKK512 pKa = 10.46LEE514 pKa = 3.84NHH516 pKa = 7.05ILNAHH521 pKa = 7.08PKK523 pKa = 9.77PIVQMACPDD532 pKa = 3.72CHH534 pKa = 8.13VMVAAFKK541 pKa = 10.14MDD543 pKa = 3.2AHH545 pKa = 7.23IKK547 pKa = 9.85NSHH550 pKa = 6.16PPSTKK555 pKa = 10.02VKK557 pKa = 10.55CSFCEE562 pKa = 4.13IQCKK566 pKa = 10.29DD567 pKa = 3.35SNRR570 pKa = 11.84LANHH574 pKa = 6.75IANVHH579 pKa = 5.46RR580 pKa = 11.84VMEE583 pKa = 4.42VKK585 pKa = 10.42PEE587 pKa = 4.04SAFSSDD593 pKa = 3.05EE594 pKa = 4.3TIKK597 pKa = 10.97VATRR601 pKa = 11.84PFLGRR606 pKa = 11.84SQRR609 pKa = 11.84PRR611 pKa = 11.84SNSPRR616 pKa = 11.84MRR618 pKa = 11.84PQQLSRR624 pKa = 11.84TSNVSTKK631 pKa = 10.03SQNSPSLEE639 pKa = 3.95AVLSLMTASLQNIEE653 pKa = 4.45KK654 pKa = 10.24RR655 pKa = 11.84LNAKK659 pKa = 10.01PPGTDD664 pKa = 3.35GQSSGIGPNN673 pKa = 3.15
Molecular weight: 74.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1047 |
374 |
673 |
523.5 |
58.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.495 ± 1.128 | 1.624 ± 0.01 |
4.871 ± 1.467 | 6.017 ± 0.205 |
3.534 ± 0.787 | 5.922 ± 0.292 |
2.865 ± 0.447 | 5.922 ± 0.116 |
5.349 ± 0.001 | 8.883 ± 0.786 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.961 ± 0.262 | 5.062 ± 0.009 |
5.158 ± 0.312 | 3.725 ± 0.009 |
5.253 ± 0.048 | 7.545 ± 0.574 |
5.158 ± 1.128 | 8.596 ± 0.973 |
1.815 ± 0.437 | 3.247 ± 0.252 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
