
Flexivirga caeni
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Dermacoccaceae; Flexivirga
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
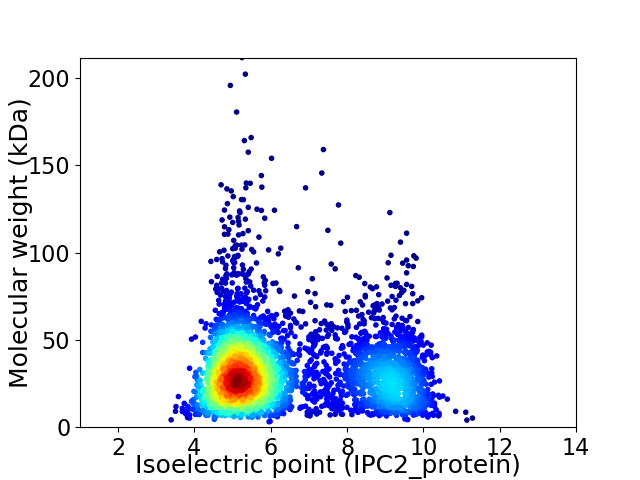
Virtual 2D-PAGE plot for 3842 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
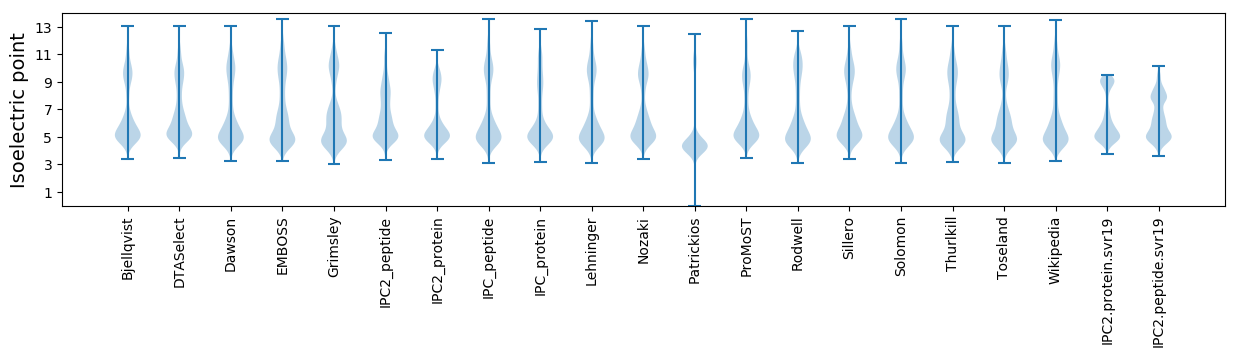
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M9MCJ1|A0A3M9MCJ1_9MICO TetR/AcrR family transcriptional regulator OS=Flexivirga caeni OX=2294115 GN=EFY87_07605 PE=4 SV=1
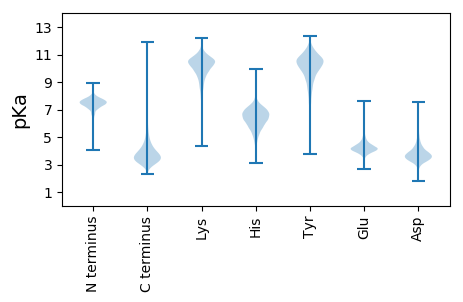
MM1 pKa = 7.9SDD3 pKa = 3.62LEE5 pKa = 4.32YY6 pKa = 10.16PADD9 pKa = 3.54LRR11 pKa = 11.84YY12 pKa = 8.63TSDD15 pKa = 3.62HH16 pKa = 6.25EE17 pKa = 4.45WVKK20 pKa = 11.23DD21 pKa = 3.5RR22 pKa = 11.84GDD24 pKa = 3.41GVVRR28 pKa = 11.84VGISAFAQDD37 pKa = 3.46ALGDD41 pKa = 3.82VVYY44 pKa = 11.12VSLPAVGDD52 pKa = 3.97SVAAGDD58 pKa = 3.5ACGEE62 pKa = 4.23VEE64 pKa = 4.37STKK67 pKa = 10.75SVSDD71 pKa = 4.02LYY73 pKa = 11.48APLAGEE79 pKa = 3.72ITAINEE85 pKa = 4.06QLDD88 pKa = 3.89ATPEE92 pKa = 4.02LVNNDD97 pKa = 3.67PYY99 pKa = 11.7GEE101 pKa = 3.78GWMYY105 pKa = 10.37EE106 pKa = 4.32LKK108 pKa = 10.87LADD111 pKa = 4.82DD112 pKa = 4.53GALAGLQDD120 pKa = 3.34VDD122 pKa = 4.04AYY124 pKa = 10.24KK125 pKa = 10.06ATLTT129 pKa = 3.95
MM1 pKa = 7.9SDD3 pKa = 3.62LEE5 pKa = 4.32YY6 pKa = 10.16PADD9 pKa = 3.54LRR11 pKa = 11.84YY12 pKa = 8.63TSDD15 pKa = 3.62HH16 pKa = 6.25EE17 pKa = 4.45WVKK20 pKa = 11.23DD21 pKa = 3.5RR22 pKa = 11.84GDD24 pKa = 3.41GVVRR28 pKa = 11.84VGISAFAQDD37 pKa = 3.46ALGDD41 pKa = 3.82VVYY44 pKa = 11.12VSLPAVGDD52 pKa = 3.97SVAAGDD58 pKa = 3.5ACGEE62 pKa = 4.23VEE64 pKa = 4.37STKK67 pKa = 10.75SVSDD71 pKa = 4.02LYY73 pKa = 11.48APLAGEE79 pKa = 3.72ITAINEE85 pKa = 4.06QLDD88 pKa = 3.89ATPEE92 pKa = 4.02LVNNDD97 pKa = 3.67PYY99 pKa = 11.7GEE101 pKa = 3.78GWMYY105 pKa = 10.37EE106 pKa = 4.32LKK108 pKa = 10.87LADD111 pKa = 4.82DD112 pKa = 4.53GALAGLQDD120 pKa = 3.34VDD122 pKa = 4.04AYY124 pKa = 10.24KK125 pKa = 10.06ATLTT129 pKa = 3.95
Molecular weight: 13.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M9M2S8|A0A3M9M2S8_9MICO SDR family NAD(P)-dependent oxidoreductase OS=Flexivirga caeni OX=2294115 GN=EFY87_15835 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.39GRR40 pKa = 11.84AKK42 pKa = 10.3ISAA45 pKa = 3.71
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.39GRR40 pKa = 11.84AKK42 pKa = 10.3ISAA45 pKa = 3.71
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1225569 |
28 |
2003 |
319.0 |
34.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.032 ± 0.048 | 0.806 ± 0.011 |
6.16 ± 0.036 | 5.116 ± 0.041 |
2.881 ± 0.024 | 8.996 ± 0.036 |
2.363 ± 0.02 | 4.069 ± 0.023 |
2.101 ± 0.029 | 10.098 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.91 ± 0.016 | 1.886 ± 0.02 |
5.536 ± 0.028 | 3.224 ± 0.022 |
7.607 ± 0.043 | 5.595 ± 0.03 |
6.192 ± 0.027 | 8.866 ± 0.035 |
1.567 ± 0.017 | 1.997 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
