
Salinispira pacifica
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Spirochaetaceae; Salinispira
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
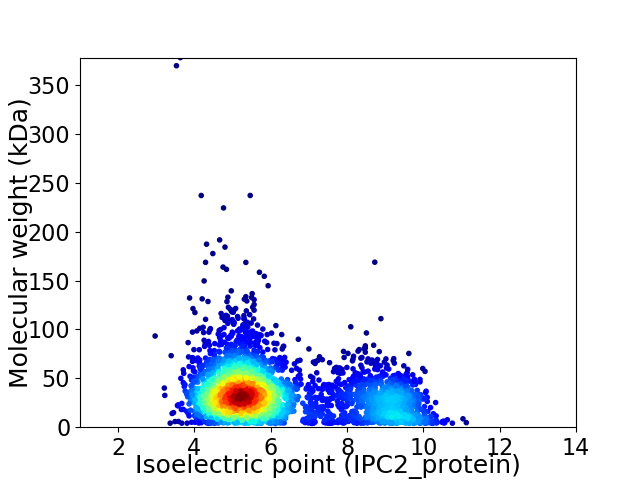
Virtual 2D-PAGE plot for 3350 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V5WCD2|V5WCD2_9SPIO Beta sliding clamp OS=Salinispira pacifica OX=1307761 GN=L21SP2_0002 PE=3 SV=1
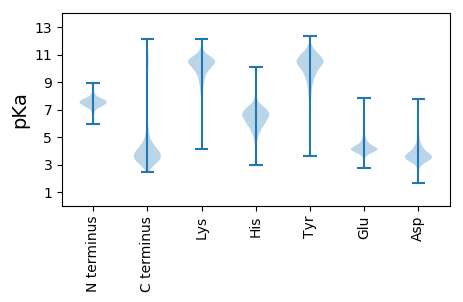
MM1 pKa = 7.66VLDD4 pKa = 3.88FRR6 pKa = 11.84EE7 pKa = 5.1GYY9 pKa = 9.74ILQQIKK15 pKa = 9.7EE16 pKa = 4.37RR17 pKa = 11.84PVKK20 pKa = 10.19QYY22 pKa = 10.73QIHH25 pKa = 5.91EE26 pKa = 4.6KK27 pKa = 10.47IFILLLIPAAFLLASCDD44 pKa = 3.42FTGIDD49 pKa = 3.22NSAVLEE55 pKa = 4.12EE56 pKa = 4.94LAGDD60 pKa = 3.53WWLTDD65 pKa = 2.9VDD67 pKa = 4.79VYY69 pKa = 11.83ADD71 pKa = 3.72GQEE74 pKa = 4.36VTNSLPDD81 pKa = 3.37TLYY84 pKa = 10.29WPDD87 pKa = 3.21SVFAAEE93 pKa = 4.05GLEE96 pKa = 4.17YY97 pKa = 9.75PDD99 pKa = 4.88SVFITDD105 pKa = 3.74KK106 pKa = 10.79GWYY109 pKa = 8.97PSDD112 pKa = 3.45AYY114 pKa = 11.07YY115 pKa = 10.39PNDD118 pKa = 3.24AYY120 pKa = 11.55YY121 pKa = 9.43PDD123 pKa = 3.8SVFYY127 pKa = 10.71FPDD130 pKa = 3.34SVFFPDD136 pKa = 4.49SSFFPDD142 pKa = 3.55SFFFPDD148 pKa = 3.6SVFFPDD154 pKa = 4.71SVFQHH159 pKa = 6.48FPTRR163 pKa = 11.84MTVSEE168 pKa = 4.68DD169 pKa = 3.19GSVVVAADD177 pKa = 3.86IMWDD181 pKa = 3.13THH183 pKa = 6.8GFDD186 pKa = 4.32GSLGEE191 pKa = 4.39PGQISSEE198 pKa = 4.09PTIEE202 pKa = 3.93TSISVTEE209 pKa = 4.64DD210 pKa = 2.83GMLQTAGQNQVFTSPRR226 pKa = 11.84SGSRR230 pKa = 11.84NSGFSNWPKK239 pKa = 10.42NYY241 pKa = 9.89EE242 pKa = 3.64ITEE245 pKa = 4.22NGDD248 pKa = 3.95LILSKK253 pKa = 9.26TQTIDD258 pKa = 3.13GVEE261 pKa = 4.22YY262 pKa = 9.84TVEE265 pKa = 3.78EE266 pKa = 4.55HH267 pKa = 5.81FTRR270 pKa = 11.84EE271 pKa = 3.99APGAPDD277 pKa = 3.76FGGVLGWWEE286 pKa = 3.96YY287 pKa = 9.38TGKK290 pKa = 10.25EE291 pKa = 3.97SGMADD296 pKa = 3.94YY297 pKa = 11.09IHH299 pKa = 7.31ISPMDD304 pKa = 3.44IASSFGLIDD313 pKa = 3.46SGQSVVTGPLAGNIILWSYY332 pKa = 11.18SEE334 pKa = 4.38SDD336 pKa = 3.48STAGPSSGDD345 pKa = 3.34KK346 pKa = 11.06LLDD349 pKa = 3.98AGGLDD354 pKa = 3.48VGYY357 pKa = 9.77NAPDD361 pKa = 3.43GEE363 pKa = 4.65MILNQLSNTSPATSSSDD380 pKa = 2.61WTLNTFTWTYY390 pKa = 10.42SISGDD395 pKa = 3.37TLTIITGSTTLTYY408 pKa = 11.02DD409 pKa = 4.64KK410 pKa = 10.85IQAQQ414 pKa = 3.5
MM1 pKa = 7.66VLDD4 pKa = 3.88FRR6 pKa = 11.84EE7 pKa = 5.1GYY9 pKa = 9.74ILQQIKK15 pKa = 9.7EE16 pKa = 4.37RR17 pKa = 11.84PVKK20 pKa = 10.19QYY22 pKa = 10.73QIHH25 pKa = 5.91EE26 pKa = 4.6KK27 pKa = 10.47IFILLLIPAAFLLASCDD44 pKa = 3.42FTGIDD49 pKa = 3.22NSAVLEE55 pKa = 4.12EE56 pKa = 4.94LAGDD60 pKa = 3.53WWLTDD65 pKa = 2.9VDD67 pKa = 4.79VYY69 pKa = 11.83ADD71 pKa = 3.72GQEE74 pKa = 4.36VTNSLPDD81 pKa = 3.37TLYY84 pKa = 10.29WPDD87 pKa = 3.21SVFAAEE93 pKa = 4.05GLEE96 pKa = 4.17YY97 pKa = 9.75PDD99 pKa = 4.88SVFITDD105 pKa = 3.74KK106 pKa = 10.79GWYY109 pKa = 8.97PSDD112 pKa = 3.45AYY114 pKa = 11.07YY115 pKa = 10.39PNDD118 pKa = 3.24AYY120 pKa = 11.55YY121 pKa = 9.43PDD123 pKa = 3.8SVFYY127 pKa = 10.71FPDD130 pKa = 3.34SVFFPDD136 pKa = 4.49SSFFPDD142 pKa = 3.55SFFFPDD148 pKa = 3.6SVFFPDD154 pKa = 4.71SVFQHH159 pKa = 6.48FPTRR163 pKa = 11.84MTVSEE168 pKa = 4.68DD169 pKa = 3.19GSVVVAADD177 pKa = 3.86IMWDD181 pKa = 3.13THH183 pKa = 6.8GFDD186 pKa = 4.32GSLGEE191 pKa = 4.39PGQISSEE198 pKa = 4.09PTIEE202 pKa = 3.93TSISVTEE209 pKa = 4.64DD210 pKa = 2.83GMLQTAGQNQVFTSPRR226 pKa = 11.84SGSRR230 pKa = 11.84NSGFSNWPKK239 pKa = 10.42NYY241 pKa = 9.89EE242 pKa = 3.64ITEE245 pKa = 4.22NGDD248 pKa = 3.95LILSKK253 pKa = 9.26TQTIDD258 pKa = 3.13GVEE261 pKa = 4.22YY262 pKa = 9.84TVEE265 pKa = 3.78EE266 pKa = 4.55HH267 pKa = 5.81FTRR270 pKa = 11.84EE271 pKa = 3.99APGAPDD277 pKa = 3.76FGGVLGWWEE286 pKa = 3.96YY287 pKa = 9.38TGKK290 pKa = 10.25EE291 pKa = 3.97SGMADD296 pKa = 3.94YY297 pKa = 11.09IHH299 pKa = 7.31ISPMDD304 pKa = 3.44IASSFGLIDD313 pKa = 3.46SGQSVVTGPLAGNIILWSYY332 pKa = 11.18SEE334 pKa = 4.38SDD336 pKa = 3.48STAGPSSGDD345 pKa = 3.34KK346 pKa = 11.06LLDD349 pKa = 3.98AGGLDD354 pKa = 3.48VGYY357 pKa = 9.77NAPDD361 pKa = 3.43GEE363 pKa = 4.65MILNQLSNTSPATSSSDD380 pKa = 2.61WTLNTFTWTYY390 pKa = 10.42SISGDD395 pKa = 3.37TLTIITGSTTLTYY408 pKa = 11.02DD409 pKa = 4.64KK410 pKa = 10.85IQAQQ414 pKa = 3.5
Molecular weight: 45.54 kDa
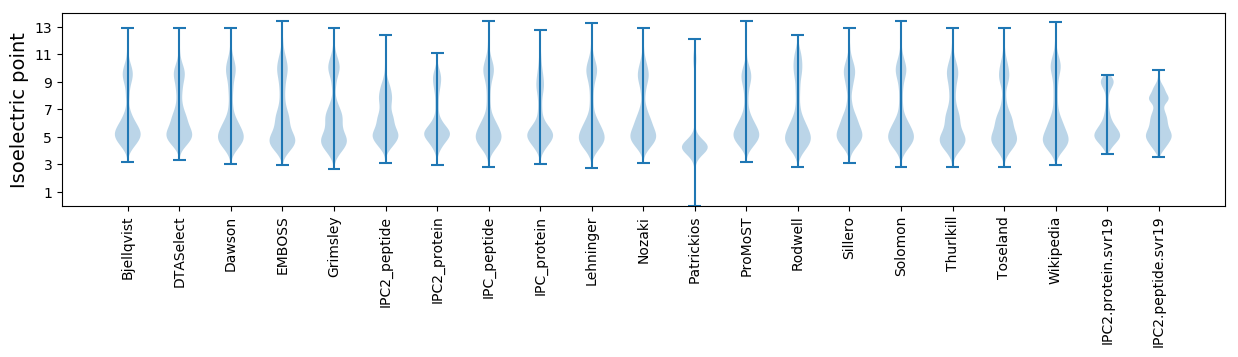
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V5WJ47|V5WJ47_9SPIO Uncharacterized protein OS=Salinispira pacifica OX=1307761 GN=L21SP2_1816 PE=4 SV=1
MM1 pKa = 7.81RR2 pKa = 11.84IVRR5 pKa = 11.84SLIGRR10 pKa = 11.84AHH12 pKa = 7.73IGRR15 pKa = 11.84PSPGNIHH22 pKa = 6.3RR23 pKa = 11.84QPVRR27 pKa = 11.84NSMLNRR33 pKa = 11.84RR34 pKa = 11.84GAPGIFRR41 pKa = 4.43
MM1 pKa = 7.81RR2 pKa = 11.84IVRR5 pKa = 11.84SLIGRR10 pKa = 11.84AHH12 pKa = 7.73IGRR15 pKa = 11.84PSPGNIHH22 pKa = 6.3RR23 pKa = 11.84QPVRR27 pKa = 11.84NSMLNRR33 pKa = 11.84RR34 pKa = 11.84GAPGIFRR41 pKa = 4.43
Molecular weight: 4.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1098157 |
37 |
3663 |
327.8 |
36.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.693 ± 0.041 | 0.665 ± 0.013 |
5.722 ± 0.044 | 7.329 ± 0.047 |
4.358 ± 0.026 | 7.758 ± 0.051 |
2.097 ± 0.022 | 6.74 ± 0.035 |
4.124 ± 0.049 | 10.269 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.589 ± 0.022 | 3.802 ± 0.032 |
4.473 ± 0.032 | 3.636 ± 0.028 |
6.464 ± 0.042 | 7.44 ± 0.036 |
4.523 ± 0.039 | 6.043 ± 0.042 |
1.081 ± 0.015 | 3.194 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
