
Lachnospiraceae bacterium NE2001
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 5.68
Get precalculated fractions of proteins
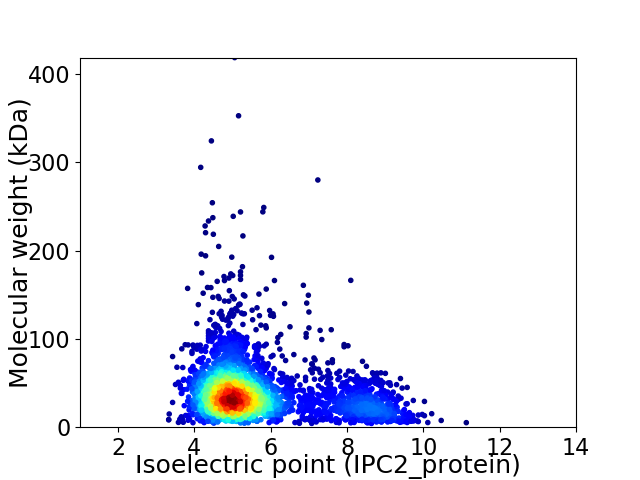
Virtual 2D-PAGE plot for 3028 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9ITX5|A0A1H9ITX5_9FIRM Uncharacterized protein OS=Lachnospiraceae bacterium NE2001 OX=1520823 GN=SAMN02910369_02409 PE=4 SV=1
MM1 pKa = 7.71KK2 pKa = 10.11FRR4 pKa = 11.84RR5 pKa = 11.84LSAVLVAVMILSLVSGCGKK24 pKa = 10.05KK25 pKa = 9.92EE26 pKa = 3.52KK27 pKa = 10.57SAIDD31 pKa = 3.69EE32 pKa = 4.33MKK34 pKa = 10.72SNYY37 pKa = 9.07IKK39 pKa = 10.93YY40 pKa = 8.37VTLGAYY46 pKa = 9.5KK47 pKa = 10.35GVEE50 pKa = 3.91YY51 pKa = 10.25TPAHH55 pKa = 5.71TEE57 pKa = 3.48ITDD60 pKa = 4.66DD61 pKa = 4.14YY62 pKa = 10.82IQYY65 pKa = 10.74DD66 pKa = 3.67IDD68 pKa = 4.25NLISQNTTEE77 pKa = 4.0NQIMEE82 pKa = 5.49GIATMGDD89 pKa = 3.19AVNIDD94 pKa = 3.47FVGYY98 pKa = 10.02IDD100 pKa = 5.84GEE102 pKa = 4.54TFDD105 pKa = 5.26GGDD108 pKa = 3.52SKK110 pKa = 11.65GAGYY114 pKa = 10.68EE115 pKa = 3.96LTLGSGSFIDD125 pKa = 5.01DD126 pKa = 4.73FEE128 pKa = 4.63DD129 pKa = 3.75QICGHH134 pKa = 6.06SPGDD138 pKa = 3.6AFDD141 pKa = 4.35VEE143 pKa = 4.96VTFPDD148 pKa = 4.1DD149 pKa = 3.97YY150 pKa = 11.78GNEE153 pKa = 3.96DD154 pKa = 4.01LAGKK158 pKa = 9.89DD159 pKa = 3.38AVFEE163 pKa = 4.32TTLNYY168 pKa = 9.8IIEE171 pKa = 4.42KK172 pKa = 10.44VEE174 pKa = 3.86PEE176 pKa = 4.12YY177 pKa = 11.51NDD179 pKa = 4.04ALVASATDD187 pKa = 3.57YY188 pKa = 9.6ATTDD192 pKa = 3.12EE193 pKa = 4.93FEE195 pKa = 4.14TAKK198 pKa = 10.7RR199 pKa = 11.84EE200 pKa = 4.2AYY202 pKa = 8.0EE203 pKa = 4.08AQAAEE208 pKa = 4.27SDD210 pKa = 3.9LANDD214 pKa = 3.55KK215 pKa = 10.03STVFNKK221 pKa = 9.98VIEE224 pKa = 4.74DD225 pKa = 3.94STVSEE230 pKa = 4.43YY231 pKa = 10.59PEE233 pKa = 4.75SEE235 pKa = 4.19VNDD238 pKa = 4.98RR239 pKa = 11.84IQMVMDD245 pKa = 4.25SVQQEE250 pKa = 4.2AEE252 pKa = 4.17ANGVDD257 pKa = 3.12IDD259 pKa = 4.71TYY261 pKa = 10.88LSNYY265 pKa = 9.92GYY267 pKa = 10.94DD268 pKa = 3.67LDD270 pKa = 4.77SFKK273 pKa = 11.62DD274 pKa = 3.63NIKK277 pKa = 9.95TSVEE281 pKa = 3.9TYY283 pKa = 9.76IRR285 pKa = 11.84EE286 pKa = 3.95KK287 pKa = 10.16MIVVAIADD295 pKa = 3.72AEE297 pKa = 4.88GITVTDD303 pKa = 3.62EE304 pKa = 4.42EE305 pKa = 4.64VDD307 pKa = 3.57QKK309 pKa = 11.27VQEE312 pKa = 4.31LLEE315 pKa = 4.26QTGLTDD321 pKa = 5.75KK322 pKa = 9.38EE323 pKa = 4.67TLSQQYY329 pKa = 9.81GFKK332 pKa = 10.96DD333 pKa = 2.85EE334 pKa = 4.58DD335 pKa = 4.0YY336 pKa = 9.79YY337 pKa = 11.98YY338 pKa = 10.76EE339 pKa = 3.99VLYY342 pKa = 11.09SKK344 pKa = 10.29IYY346 pKa = 10.7DD347 pKa = 4.56FIYY350 pKa = 9.91EE351 pKa = 3.89NAVAVEE357 pKa = 4.25ASNTDD362 pKa = 3.29ADD364 pKa = 4.24EE365 pKa = 4.77DD366 pKa = 4.49ASDD369 pKa = 4.35SDD371 pKa = 4.86GLVDD375 pKa = 5.77DD376 pKa = 4.8YY377 pKa = 11.64GTTEE381 pKa = 3.94EE382 pKa = 4.41EE383 pKa = 4.17
MM1 pKa = 7.71KK2 pKa = 10.11FRR4 pKa = 11.84RR5 pKa = 11.84LSAVLVAVMILSLVSGCGKK24 pKa = 10.05KK25 pKa = 9.92EE26 pKa = 3.52KK27 pKa = 10.57SAIDD31 pKa = 3.69EE32 pKa = 4.33MKK34 pKa = 10.72SNYY37 pKa = 9.07IKK39 pKa = 10.93YY40 pKa = 8.37VTLGAYY46 pKa = 9.5KK47 pKa = 10.35GVEE50 pKa = 3.91YY51 pKa = 10.25TPAHH55 pKa = 5.71TEE57 pKa = 3.48ITDD60 pKa = 4.66DD61 pKa = 4.14YY62 pKa = 10.82IQYY65 pKa = 10.74DD66 pKa = 3.67IDD68 pKa = 4.25NLISQNTTEE77 pKa = 4.0NQIMEE82 pKa = 5.49GIATMGDD89 pKa = 3.19AVNIDD94 pKa = 3.47FVGYY98 pKa = 10.02IDD100 pKa = 5.84GEE102 pKa = 4.54TFDD105 pKa = 5.26GGDD108 pKa = 3.52SKK110 pKa = 11.65GAGYY114 pKa = 10.68EE115 pKa = 3.96LTLGSGSFIDD125 pKa = 5.01DD126 pKa = 4.73FEE128 pKa = 4.63DD129 pKa = 3.75QICGHH134 pKa = 6.06SPGDD138 pKa = 3.6AFDD141 pKa = 4.35VEE143 pKa = 4.96VTFPDD148 pKa = 4.1DD149 pKa = 3.97YY150 pKa = 11.78GNEE153 pKa = 3.96DD154 pKa = 4.01LAGKK158 pKa = 9.89DD159 pKa = 3.38AVFEE163 pKa = 4.32TTLNYY168 pKa = 9.8IIEE171 pKa = 4.42KK172 pKa = 10.44VEE174 pKa = 3.86PEE176 pKa = 4.12YY177 pKa = 11.51NDD179 pKa = 4.04ALVASATDD187 pKa = 3.57YY188 pKa = 9.6ATTDD192 pKa = 3.12EE193 pKa = 4.93FEE195 pKa = 4.14TAKK198 pKa = 10.7RR199 pKa = 11.84EE200 pKa = 4.2AYY202 pKa = 8.0EE203 pKa = 4.08AQAAEE208 pKa = 4.27SDD210 pKa = 3.9LANDD214 pKa = 3.55KK215 pKa = 10.03STVFNKK221 pKa = 9.98VIEE224 pKa = 4.74DD225 pKa = 3.94STVSEE230 pKa = 4.43YY231 pKa = 10.59PEE233 pKa = 4.75SEE235 pKa = 4.19VNDD238 pKa = 4.98RR239 pKa = 11.84IQMVMDD245 pKa = 4.25SVQQEE250 pKa = 4.2AEE252 pKa = 4.17ANGVDD257 pKa = 3.12IDD259 pKa = 4.71TYY261 pKa = 10.88LSNYY265 pKa = 9.92GYY267 pKa = 10.94DD268 pKa = 3.67LDD270 pKa = 4.77SFKK273 pKa = 11.62DD274 pKa = 3.63NIKK277 pKa = 9.95TSVEE281 pKa = 3.9TYY283 pKa = 9.76IRR285 pKa = 11.84EE286 pKa = 3.95KK287 pKa = 10.16MIVVAIADD295 pKa = 3.72AEE297 pKa = 4.88GITVTDD303 pKa = 3.62EE304 pKa = 4.42EE305 pKa = 4.64VDD307 pKa = 3.57QKK309 pKa = 11.27VQEE312 pKa = 4.31LLEE315 pKa = 4.26QTGLTDD321 pKa = 5.75KK322 pKa = 9.38EE323 pKa = 4.67TLSQQYY329 pKa = 9.81GFKK332 pKa = 10.96DD333 pKa = 2.85EE334 pKa = 4.58DD335 pKa = 4.0YY336 pKa = 9.79YY337 pKa = 11.98YY338 pKa = 10.76EE339 pKa = 3.99VLYY342 pKa = 11.09SKK344 pKa = 10.29IYY346 pKa = 10.7DD347 pKa = 4.56FIYY350 pKa = 9.91EE351 pKa = 3.89NAVAVEE357 pKa = 4.25ASNTDD362 pKa = 3.29ADD364 pKa = 4.24EE365 pKa = 4.77DD366 pKa = 4.49ASDD369 pKa = 4.35SDD371 pKa = 4.86GLVDD375 pKa = 5.77DD376 pKa = 4.8YY377 pKa = 11.64GTTEE381 pKa = 3.94EE382 pKa = 4.41EE383 pKa = 4.17
Molecular weight: 42.55 kDa
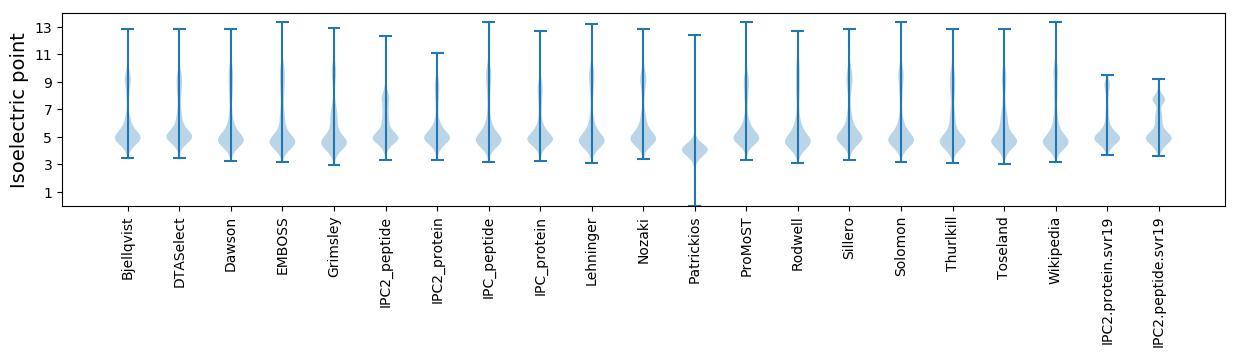
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H8YSW5|A0A1H8YSW5_9FIRM Transglutaminase-like superfamily protein OS=Lachnospiraceae bacterium NE2001 OX=1520823 GN=SAMN02910369_00106 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.74VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.03GRR39 pKa = 11.84KK40 pKa = 8.97SLTVV44 pKa = 3.12
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.74VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.03GRR39 pKa = 11.84KK40 pKa = 8.97SLTVV44 pKa = 3.12
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1117602 |
40 |
3668 |
369.1 |
41.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.799 ± 0.045 | 1.193 ± 0.017 |
6.91 ± 0.035 | 7.688 ± 0.051 |
4.177 ± 0.03 | 6.774 ± 0.038 |
1.458 ± 0.017 | 7.793 ± 0.044 |
7.233 ± 0.054 | 8.328 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.99 ± 0.027 | 4.906 ± 0.042 |
3.051 ± 0.027 | 2.541 ± 0.025 |
4.045 ± 0.034 | 6.413 ± 0.04 |
5.506 ± 0.053 | 6.757 ± 0.034 |
0.856 ± 0.018 | 4.583 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
