
Nocardiopsis sp. L17-MgMaSL7
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Nocardiopsis; unclassified Nocardiopsis
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
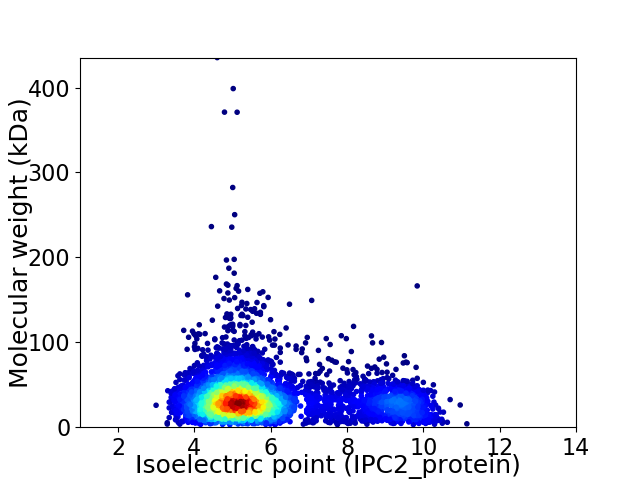
Virtual 2D-PAGE plot for 5387 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
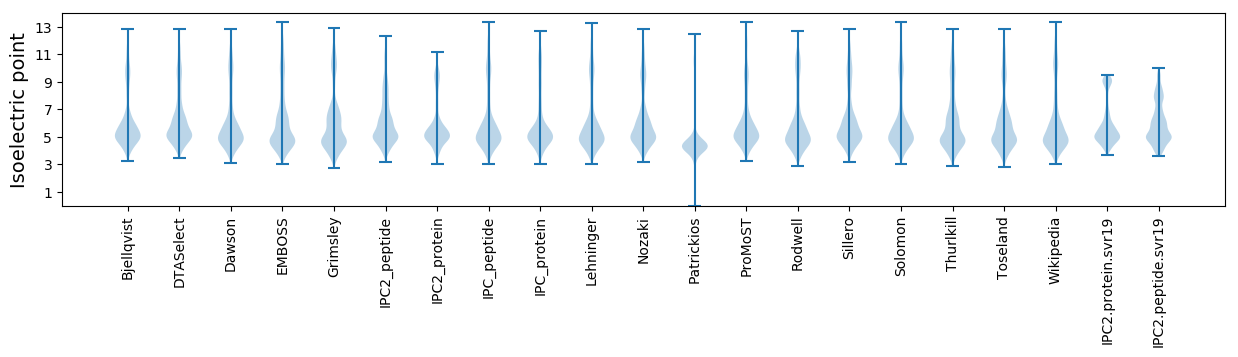
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317M2F9|A0A317M2F9_9ACTN Putative phenylalanine aminotransferase OS=Nocardiopsis sp. L17-MgMaSL7 OX=1938893 GN=pat PE=3 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84SLRR5 pKa = 11.84LPAALAALVLVASACSGEE23 pKa = 4.3GAGGDD28 pKa = 3.26SGGYY32 pKa = 8.54PRR34 pKa = 11.84NEE36 pKa = 3.95TLYY39 pKa = 8.46TTGTAWGPPTNWNPIMEE56 pKa = 4.57GNYY59 pKa = 10.45AVGTVGLVYY68 pKa = 10.58EE69 pKa = 4.56PLFVFDD75 pKa = 4.85PSEE78 pKa = 3.91GAYY81 pKa = 9.12TPWLAEE87 pKa = 3.79SDD89 pKa = 3.69EE90 pKa = 4.36WPDD93 pKa = 3.41EE94 pKa = 4.11NTHH97 pKa = 6.67VITLRR102 pKa = 11.84DD103 pKa = 3.59GVEE106 pKa = 3.96WTDD109 pKa = 3.74GEE111 pKa = 4.4PLVAQDD117 pKa = 3.88VVTTLEE123 pKa = 4.37LGHH126 pKa = 6.69FDD128 pKa = 2.79IHH130 pKa = 7.27YY131 pKa = 10.35SNVWDD136 pKa = 3.74YY137 pKa = 11.86LEE139 pKa = 4.36SAEE142 pKa = 4.15ATGEE146 pKa = 4.17RR147 pKa = 11.84EE148 pKa = 4.03VTLTFSDD155 pKa = 4.06PRR157 pKa = 11.84PQEE160 pKa = 3.54WMNWVANRR168 pKa = 11.84AIVPDD173 pKa = 5.19HH174 pKa = 6.19IWADD178 pKa = 3.45KK179 pKa = 10.97SQEE182 pKa = 4.05QVNDD186 pKa = 3.6DD187 pKa = 4.05PNRR190 pKa = 11.84DD191 pKa = 3.56PVGTGAYY198 pKa = 9.74VYY200 pKa = 10.1EE201 pKa = 4.16NHH203 pKa = 7.25ADD205 pKa = 3.87DD206 pKa = 4.36RR207 pKa = 11.84MVWRR211 pKa = 11.84RR212 pKa = 11.84NDD214 pKa = 3.45DD215 pKa = 2.82WWGTEE220 pKa = 4.0ALDD223 pKa = 5.38LEE225 pKa = 4.68MKK227 pKa = 8.39PTYY230 pKa = 10.25IVDD233 pKa = 3.38IVNASNDD240 pKa = 3.05VTMNMLNNHH249 pKa = 5.79EE250 pKa = 4.48VDD252 pKa = 3.68LSNNFLPGIDD262 pKa = 3.38QVISNDD268 pKa = 3.53NNITSYY274 pKa = 10.73FAGPPYY280 pKa = 9.52MQSANTAWLVPNHH293 pKa = 5.85EE294 pKa = 4.71HH295 pKa = 6.98PALGDD300 pKa = 3.5PEE302 pKa = 4.18FRR304 pKa = 11.84RR305 pKa = 11.84ALAHH309 pKa = 7.15AVDD312 pKa = 4.32TSQIVEE318 pKa = 4.23GPYY321 pKa = 9.98TNLVEE326 pKa = 4.75AADD329 pKa = 4.01PTGLLPQWEE338 pKa = 4.62EE339 pKa = 4.31YY340 pKa = 9.9IDD342 pKa = 4.08HH343 pKa = 6.88EE344 pKa = 4.53LVADD348 pKa = 3.88EE349 pKa = 4.91GFTYY353 pKa = 10.71DD354 pKa = 3.28SGEE357 pKa = 3.81AVAILEE363 pKa = 4.2AAGYY367 pKa = 10.3LDD369 pKa = 4.02EE370 pKa = 7.35DD371 pKa = 4.02GDD373 pKa = 4.52GFVEE377 pKa = 4.7TPDD380 pKa = 4.14GEE382 pKa = 5.06PITLSLEE389 pKa = 4.45VPSGWSDD396 pKa = 2.52WMEE399 pKa = 4.05AANVIAEE406 pKa = 4.21NAQEE410 pKa = 4.02VGIKK414 pKa = 9.98VEE416 pKa = 3.99PAFPEE421 pKa = 4.17HH422 pKa = 6.8EE423 pKa = 4.02LLIEE427 pKa = 4.13NFRR430 pKa = 11.84KK431 pKa = 10.33AEE433 pKa = 3.95FDD435 pKa = 4.01LMINNDD441 pKa = 3.48RR442 pKa = 11.84SLSNTPWTYY451 pKa = 11.36YY452 pKa = 11.05DD453 pKa = 5.79FIFQLPIRR461 pKa = 11.84DD462 pKa = 3.67QQTTANYY469 pKa = 9.95ARR471 pKa = 11.84YY472 pKa = 8.89EE473 pKa = 4.01NEE475 pKa = 4.14EE476 pKa = 3.75AWEE479 pKa = 4.01LTEE482 pKa = 5.27QLAATPAEE490 pKa = 4.51DD491 pKa = 4.18AEE493 pKa = 4.48GMAAITSDD501 pKa = 3.5LQGIFLDD508 pKa = 4.62DD509 pKa = 4.47LPIIPLWYY517 pKa = 10.35NGMWSQASNQVWTNWPSEE535 pKa = 4.24DD536 pKa = 4.4EE537 pKa = 4.18GTPDD541 pKa = 4.72NFATMWNNMYY551 pKa = 10.99QMGAIYY557 pKa = 9.42TLAEE561 pKa = 4.05IEE563 pKa = 4.11LAEE566 pKa = 4.09
MM1 pKa = 7.97RR2 pKa = 11.84SLRR5 pKa = 11.84LPAALAALVLVASACSGEE23 pKa = 4.3GAGGDD28 pKa = 3.26SGGYY32 pKa = 8.54PRR34 pKa = 11.84NEE36 pKa = 3.95TLYY39 pKa = 8.46TTGTAWGPPTNWNPIMEE56 pKa = 4.57GNYY59 pKa = 10.45AVGTVGLVYY68 pKa = 10.58EE69 pKa = 4.56PLFVFDD75 pKa = 4.85PSEE78 pKa = 3.91GAYY81 pKa = 9.12TPWLAEE87 pKa = 3.79SDD89 pKa = 3.69EE90 pKa = 4.36WPDD93 pKa = 3.41EE94 pKa = 4.11NTHH97 pKa = 6.67VITLRR102 pKa = 11.84DD103 pKa = 3.59GVEE106 pKa = 3.96WTDD109 pKa = 3.74GEE111 pKa = 4.4PLVAQDD117 pKa = 3.88VVTTLEE123 pKa = 4.37LGHH126 pKa = 6.69FDD128 pKa = 2.79IHH130 pKa = 7.27YY131 pKa = 10.35SNVWDD136 pKa = 3.74YY137 pKa = 11.86LEE139 pKa = 4.36SAEE142 pKa = 4.15ATGEE146 pKa = 4.17RR147 pKa = 11.84EE148 pKa = 4.03VTLTFSDD155 pKa = 4.06PRR157 pKa = 11.84PQEE160 pKa = 3.54WMNWVANRR168 pKa = 11.84AIVPDD173 pKa = 5.19HH174 pKa = 6.19IWADD178 pKa = 3.45KK179 pKa = 10.97SQEE182 pKa = 4.05QVNDD186 pKa = 3.6DD187 pKa = 4.05PNRR190 pKa = 11.84DD191 pKa = 3.56PVGTGAYY198 pKa = 9.74VYY200 pKa = 10.1EE201 pKa = 4.16NHH203 pKa = 7.25ADD205 pKa = 3.87DD206 pKa = 4.36RR207 pKa = 11.84MVWRR211 pKa = 11.84RR212 pKa = 11.84NDD214 pKa = 3.45DD215 pKa = 2.82WWGTEE220 pKa = 4.0ALDD223 pKa = 5.38LEE225 pKa = 4.68MKK227 pKa = 8.39PTYY230 pKa = 10.25IVDD233 pKa = 3.38IVNASNDD240 pKa = 3.05VTMNMLNNHH249 pKa = 5.79EE250 pKa = 4.48VDD252 pKa = 3.68LSNNFLPGIDD262 pKa = 3.38QVISNDD268 pKa = 3.53NNITSYY274 pKa = 10.73FAGPPYY280 pKa = 9.52MQSANTAWLVPNHH293 pKa = 5.85EE294 pKa = 4.71HH295 pKa = 6.98PALGDD300 pKa = 3.5PEE302 pKa = 4.18FRR304 pKa = 11.84RR305 pKa = 11.84ALAHH309 pKa = 7.15AVDD312 pKa = 4.32TSQIVEE318 pKa = 4.23GPYY321 pKa = 9.98TNLVEE326 pKa = 4.75AADD329 pKa = 4.01PTGLLPQWEE338 pKa = 4.62EE339 pKa = 4.31YY340 pKa = 9.9IDD342 pKa = 4.08HH343 pKa = 6.88EE344 pKa = 4.53LVADD348 pKa = 3.88EE349 pKa = 4.91GFTYY353 pKa = 10.71DD354 pKa = 3.28SGEE357 pKa = 3.81AVAILEE363 pKa = 4.2AAGYY367 pKa = 10.3LDD369 pKa = 4.02EE370 pKa = 7.35DD371 pKa = 4.02GDD373 pKa = 4.52GFVEE377 pKa = 4.7TPDD380 pKa = 4.14GEE382 pKa = 5.06PITLSLEE389 pKa = 4.45VPSGWSDD396 pKa = 2.52WMEE399 pKa = 4.05AANVIAEE406 pKa = 4.21NAQEE410 pKa = 4.02VGIKK414 pKa = 9.98VEE416 pKa = 3.99PAFPEE421 pKa = 4.17HH422 pKa = 6.8EE423 pKa = 4.02LLIEE427 pKa = 4.13NFRR430 pKa = 11.84KK431 pKa = 10.33AEE433 pKa = 3.95FDD435 pKa = 4.01LMINNDD441 pKa = 3.48RR442 pKa = 11.84SLSNTPWTYY451 pKa = 11.36YY452 pKa = 11.05DD453 pKa = 5.79FIFQLPIRR461 pKa = 11.84DD462 pKa = 3.67QQTTANYY469 pKa = 9.95ARR471 pKa = 11.84YY472 pKa = 8.89EE473 pKa = 4.01NEE475 pKa = 4.14EE476 pKa = 3.75AWEE479 pKa = 4.01LTEE482 pKa = 5.27QLAATPAEE490 pKa = 4.51DD491 pKa = 4.18AEE493 pKa = 4.48GMAAITSDD501 pKa = 3.5LQGIFLDD508 pKa = 4.62DD509 pKa = 4.47LPIIPLWYY517 pKa = 10.35NGMWSQASNQVWTNWPSEE535 pKa = 4.24DD536 pKa = 4.4EE537 pKa = 4.18GTPDD541 pKa = 4.72NFATMWNNMYY551 pKa = 10.99QMGAIYY557 pKa = 9.42TLAEE561 pKa = 4.05IEE563 pKa = 4.11LAEE566 pKa = 4.09
Molecular weight: 63.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317MEQ0|A0A317MEQ0_9ACTN CP family cyanate transporter-like MFS transporter OS=Nocardiopsis sp. L17-MgMaSL7 OX=1938893 GN=BDW27_104267 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.77LLKK22 pKa = 7.46RR23 pKa = 11.84TRR25 pKa = 11.84VARR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.94KK32 pKa = 9.77
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.77LLKK22 pKa = 7.46RR23 pKa = 11.84TRR25 pKa = 11.84VARR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.94KK32 pKa = 9.77
Molecular weight: 3.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1783097 |
27 |
4124 |
331.0 |
35.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.269 ± 0.044 | 0.695 ± 0.009 |
6.26 ± 0.028 | 6.618 ± 0.038 |
2.813 ± 0.019 | 9.462 ± 0.035 |
2.342 ± 0.018 | 3.135 ± 0.023 |
1.539 ± 0.021 | 10.413 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.88 ± 0.013 | 1.864 ± 0.019 |
6.088 ± 0.032 | 2.59 ± 0.02 |
8.154 ± 0.04 | 5.488 ± 0.024 |
5.954 ± 0.022 | 9.035 ± 0.033 |
1.492 ± 0.016 | 1.912 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
