
Leuconostoc fallax
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli;
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
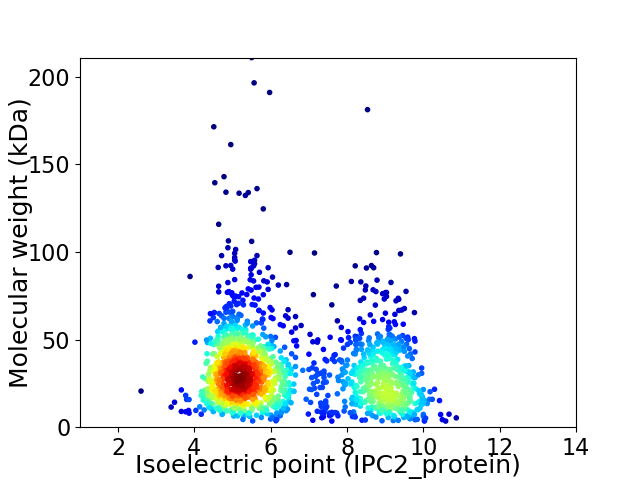
Virtual 2D-PAGE plot for 1619 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R5N8V2|A0A4R5N8V2_9LACO DUF2179 domain-containing protein OS=Leuconostoc fallax OX=1251 GN=C5L23_000613 PE=4 SV=1
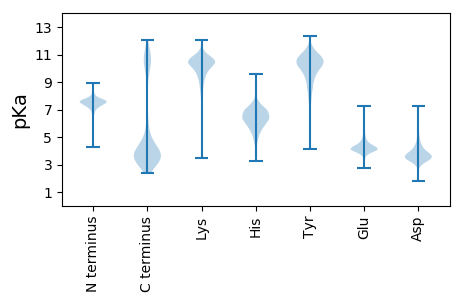
MM1 pKa = 7.22TAINDD6 pKa = 3.89ANSMLNAIEE15 pKa = 4.89SGDD18 pKa = 3.69LVSAQNYY25 pKa = 7.38FKK27 pKa = 11.04KK28 pKa = 10.59SLQNDD33 pKa = 3.61SDD35 pKa = 3.74EE36 pKa = 5.41LIYY39 pKa = 11.21NLAEE43 pKa = 3.96EE44 pKa = 4.89LYY46 pKa = 11.28GMGFTHH52 pKa = 6.34YY53 pKa = 10.68AKK55 pKa = 10.32EE56 pKa = 4.09AYY58 pKa = 9.0EE59 pKa = 4.19LLLTRR64 pKa = 11.84YY65 pKa = 8.26PQEE68 pKa = 4.05DD69 pKa = 4.11SIRR72 pKa = 11.84TALADD77 pKa = 3.32IAIDD81 pKa = 4.62DD82 pKa = 4.43GEE84 pKa = 4.56IDD86 pKa = 3.58HH87 pKa = 7.05ALEE90 pKa = 5.06LLSQVNPEE98 pKa = 3.39SDD100 pKa = 3.75AYY102 pKa = 11.14VSSLLVKK109 pKa = 10.49ADD111 pKa = 4.53LYY113 pKa = 11.12QSEE116 pKa = 4.64GLNEE120 pKa = 3.93AAEE123 pKa = 4.3SSLRR127 pKa = 11.84RR128 pKa = 11.84AQQLMPDD135 pKa = 3.4HH136 pKa = 7.55DD137 pKa = 5.17IIQLALAEE145 pKa = 4.84FYY147 pKa = 11.13FDD149 pKa = 2.9IQSYY153 pKa = 8.76HH154 pKa = 4.75QAIPIYY160 pKa = 10.06RR161 pKa = 11.84RR162 pKa = 11.84LIMQGTLEE170 pKa = 4.01VSKK173 pKa = 10.72IDD175 pKa = 3.12IVARR179 pKa = 11.84LGVAYY184 pKa = 9.88AQIGNYY190 pKa = 10.11DD191 pKa = 3.39NAIGYY196 pKa = 9.29LEE198 pKa = 4.06QIKK201 pKa = 10.4PEE203 pKa = 4.51NITLTTQFQLAVVYY217 pKa = 8.09YY218 pKa = 10.77QNTQWQDD225 pKa = 3.33AIDD228 pKa = 4.49LFNDD232 pKa = 4.64IIDD235 pKa = 3.84VDD237 pKa = 3.87PKK239 pKa = 9.18YY240 pKa = 9.54TSVYY244 pKa = 9.25PLLGKK249 pKa = 10.3AYY251 pKa = 9.31EE252 pKa = 4.24KK253 pKa = 10.93LNKK256 pKa = 9.03IDD258 pKa = 3.35EE259 pKa = 4.75AYY261 pKa = 10.43RR262 pKa = 11.84IYY264 pKa = 10.67QEE266 pKa = 5.15GLSQDD271 pKa = 3.51EE272 pKa = 4.85TNSQLYY278 pKa = 9.63RR279 pKa = 11.84LAGITAQKK287 pKa = 10.69LNEE290 pKa = 4.24LEE292 pKa = 4.11SAEE295 pKa = 4.33TYY297 pKa = 9.79LVKK300 pKa = 10.8AIALDD305 pKa = 3.96EE306 pKa = 4.89ADD308 pKa = 3.47TMAITSLAQVLLLQDD323 pKa = 3.67RR324 pKa = 11.84AAEE327 pKa = 4.34VITLILDD334 pKa = 3.45QQEE337 pKa = 3.99NDD339 pKa = 3.65VIDD342 pKa = 3.78NSFYY346 pKa = 10.15WVLAKK351 pKa = 10.33AYY353 pKa = 9.75EE354 pKa = 4.13MTDD357 pKa = 3.35EE358 pKa = 4.23YY359 pKa = 11.4EE360 pKa = 4.34KK361 pKa = 11.14ASEE364 pKa = 4.37NYY366 pKa = 10.06DD367 pKa = 3.85LAWPLLQNNTDD378 pKa = 3.9FLLDD382 pKa = 3.3AMAWYY387 pKa = 10.17RR388 pKa = 11.84EE389 pKa = 3.85NGQRR393 pKa = 11.84DD394 pKa = 3.52IEE396 pKa = 4.28IQSLQQYY403 pKa = 9.93LKK405 pKa = 10.71LVPDD409 pKa = 5.26DD410 pKa = 5.02IEE412 pKa = 4.24MQLRR416 pKa = 11.84LADD419 pKa = 3.95YY420 pKa = 10.58EE421 pKa = 4.16NDD423 pKa = 3.51SMM425 pKa = 5.97
MM1 pKa = 7.22TAINDD6 pKa = 3.89ANSMLNAIEE15 pKa = 4.89SGDD18 pKa = 3.69LVSAQNYY25 pKa = 7.38FKK27 pKa = 11.04KK28 pKa = 10.59SLQNDD33 pKa = 3.61SDD35 pKa = 3.74EE36 pKa = 5.41LIYY39 pKa = 11.21NLAEE43 pKa = 3.96EE44 pKa = 4.89LYY46 pKa = 11.28GMGFTHH52 pKa = 6.34YY53 pKa = 10.68AKK55 pKa = 10.32EE56 pKa = 4.09AYY58 pKa = 9.0EE59 pKa = 4.19LLLTRR64 pKa = 11.84YY65 pKa = 8.26PQEE68 pKa = 4.05DD69 pKa = 4.11SIRR72 pKa = 11.84TALADD77 pKa = 3.32IAIDD81 pKa = 4.62DD82 pKa = 4.43GEE84 pKa = 4.56IDD86 pKa = 3.58HH87 pKa = 7.05ALEE90 pKa = 5.06LLSQVNPEE98 pKa = 3.39SDD100 pKa = 3.75AYY102 pKa = 11.14VSSLLVKK109 pKa = 10.49ADD111 pKa = 4.53LYY113 pKa = 11.12QSEE116 pKa = 4.64GLNEE120 pKa = 3.93AAEE123 pKa = 4.3SSLRR127 pKa = 11.84RR128 pKa = 11.84AQQLMPDD135 pKa = 3.4HH136 pKa = 7.55DD137 pKa = 5.17IIQLALAEE145 pKa = 4.84FYY147 pKa = 11.13FDD149 pKa = 2.9IQSYY153 pKa = 8.76HH154 pKa = 4.75QAIPIYY160 pKa = 10.06RR161 pKa = 11.84RR162 pKa = 11.84LIMQGTLEE170 pKa = 4.01VSKK173 pKa = 10.72IDD175 pKa = 3.12IVARR179 pKa = 11.84LGVAYY184 pKa = 9.88AQIGNYY190 pKa = 10.11DD191 pKa = 3.39NAIGYY196 pKa = 9.29LEE198 pKa = 4.06QIKK201 pKa = 10.4PEE203 pKa = 4.51NITLTTQFQLAVVYY217 pKa = 8.09YY218 pKa = 10.77QNTQWQDD225 pKa = 3.33AIDD228 pKa = 4.49LFNDD232 pKa = 4.64IIDD235 pKa = 3.84VDD237 pKa = 3.87PKK239 pKa = 9.18YY240 pKa = 9.54TSVYY244 pKa = 9.25PLLGKK249 pKa = 10.3AYY251 pKa = 9.31EE252 pKa = 4.24KK253 pKa = 10.93LNKK256 pKa = 9.03IDD258 pKa = 3.35EE259 pKa = 4.75AYY261 pKa = 10.43RR262 pKa = 11.84IYY264 pKa = 10.67QEE266 pKa = 5.15GLSQDD271 pKa = 3.51EE272 pKa = 4.85TNSQLYY278 pKa = 9.63RR279 pKa = 11.84LAGITAQKK287 pKa = 10.69LNEE290 pKa = 4.24LEE292 pKa = 4.11SAEE295 pKa = 4.33TYY297 pKa = 9.79LVKK300 pKa = 10.8AIALDD305 pKa = 3.96EE306 pKa = 4.89ADD308 pKa = 3.47TMAITSLAQVLLLQDD323 pKa = 3.67RR324 pKa = 11.84AAEE327 pKa = 4.34VITLILDD334 pKa = 3.45QQEE337 pKa = 3.99NDD339 pKa = 3.65VIDD342 pKa = 3.78NSFYY346 pKa = 10.15WVLAKK351 pKa = 10.33AYY353 pKa = 9.75EE354 pKa = 4.13MTDD357 pKa = 3.35EE358 pKa = 4.23YY359 pKa = 11.4EE360 pKa = 4.34KK361 pKa = 11.14ASEE364 pKa = 4.37NYY366 pKa = 10.06DD367 pKa = 3.85LAWPLLQNNTDD378 pKa = 3.9FLLDD382 pKa = 3.3AMAWYY387 pKa = 10.17RR388 pKa = 11.84EE389 pKa = 3.85NGQRR393 pKa = 11.84DD394 pKa = 3.52IEE396 pKa = 4.28IQSLQQYY403 pKa = 9.93LKK405 pKa = 10.71LVPDD409 pKa = 5.26DD410 pKa = 5.02IEE412 pKa = 4.24MQLRR416 pKa = 11.84LADD419 pKa = 3.95YY420 pKa = 10.58EE421 pKa = 4.16NDD423 pKa = 3.51SMM425 pKa = 5.97
Molecular weight: 48.56 kDa
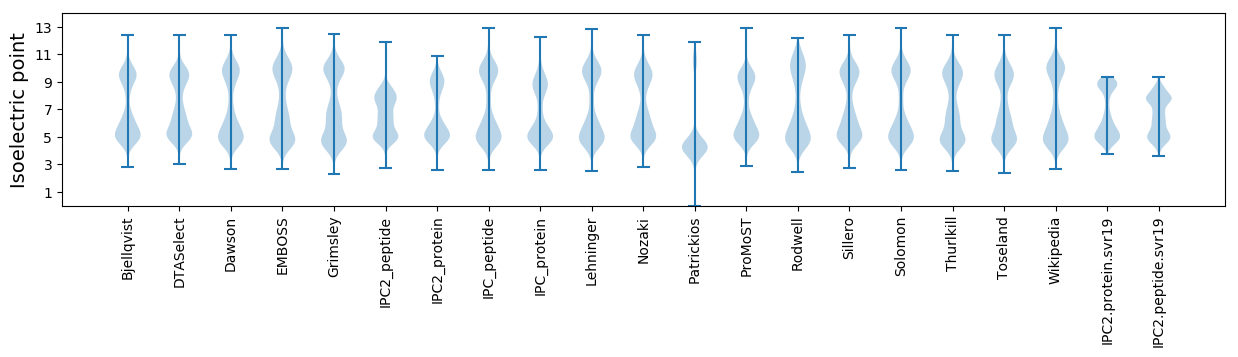
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V3A2H1|A0A4V3A2H1_9LACO Uncharacterized protein OS=Leuconostoc fallax OX=1251 GN=C5L23_000634 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.34KK9 pKa = 7.47RR10 pKa = 11.84HH11 pKa = 5.71RR12 pKa = 11.84EE13 pKa = 3.7RR14 pKa = 11.84VHH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.23VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.34KK9 pKa = 7.47RR10 pKa = 11.84HH11 pKa = 5.71RR12 pKa = 11.84EE13 pKa = 3.7RR14 pKa = 11.84VHH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.23VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
490441 |
30 |
1901 |
302.9 |
33.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.157 ± 0.07 | 0.265 ± 0.025 |
5.821 ± 0.059 | 5.235 ± 0.061 |
4.265 ± 0.051 | 6.386 ± 0.053 |
2.272 ± 0.029 | 7.735 ± 0.065 |
5.855 ± 0.059 | 9.9 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.674 ± 0.03 | 4.962 ± 0.046 |
3.399 ± 0.029 | 5.229 ± 0.06 |
4.008 ± 0.049 | 6.027 ± 0.072 |
6.208 ± 0.049 | 7.056 ± 0.049 |
1.049 ± 0.026 | 3.472 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
