
Canine mastadenovirus A
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Mastadenovirus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
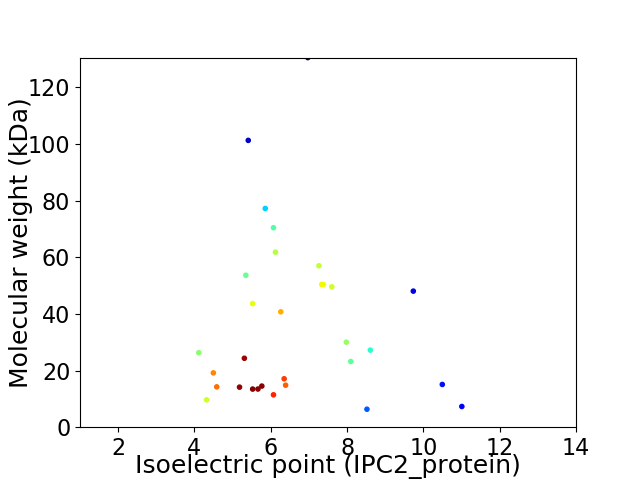
Virtual 2D-PAGE plot for 31 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
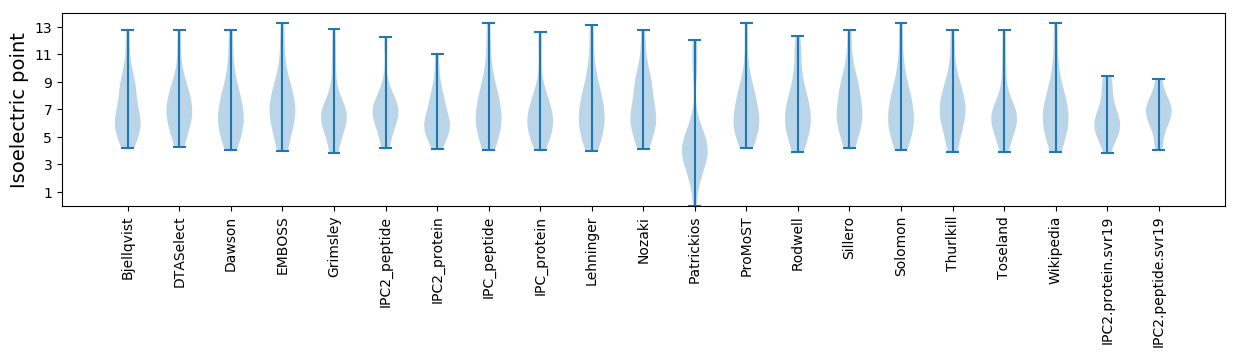
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6S6MM40|A0A6S6MM40_9ADEN E4 ORF A OS=Canine mastadenovirus A OX=10537 PE=4 SV=1
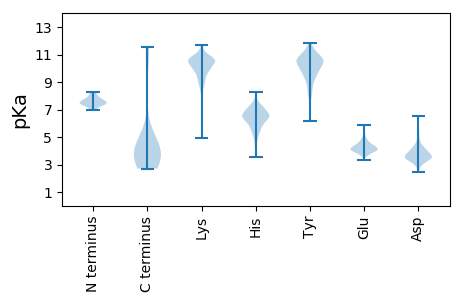
MM1 pKa = 7.96KK2 pKa = 9.44YY3 pKa = 9.12TIVPAPRR10 pKa = 11.84NLHH13 pKa = 6.86DD14 pKa = 4.27YY15 pKa = 10.64VLEE18 pKa = 4.72LLEE21 pKa = 4.54EE22 pKa = 4.15WQPDD26 pKa = 4.08CLDD29 pKa = 3.45CEE31 pKa = 4.57YY32 pKa = 10.51PHH34 pKa = 7.12GSPSPPTLHH43 pKa = 7.48DD44 pKa = 4.17LFDD47 pKa = 4.14VEE49 pKa = 5.88LEE51 pKa = 4.34TSHH54 pKa = 7.55SPFVGLCDD62 pKa = 3.43SCAEE66 pKa = 4.27ADD68 pKa = 3.78TDD70 pKa = 4.26SSASTEE76 pKa = 4.03ADD78 pKa = 3.22SGFSPLSTPPVSPIPPHH95 pKa = 5.76PTSPASISDD104 pKa = 3.82DD105 pKa = 3.14MLLCLEE111 pKa = 4.63EE112 pKa = 4.67MPTFDD117 pKa = 6.17DD118 pKa = 3.65EE119 pKa = 5.53DD120 pKa = 4.02EE121 pKa = 4.35VRR123 pKa = 11.84SAATTFEE130 pKa = 3.86RR131 pKa = 11.84WEE133 pKa = 4.09NTFDD137 pKa = 3.56PHH139 pKa = 6.94VGPIFGCLRR148 pKa = 11.84CAFYY152 pKa = 10.82QEE154 pKa = 4.06QDD156 pKa = 3.54DD157 pKa = 4.13NALCGLCYY165 pKa = 10.43LKK167 pKa = 10.94ALAEE171 pKa = 4.38VPFALPVRR179 pKa = 11.84SEE181 pKa = 3.97PASAGAEE188 pKa = 4.06EE189 pKa = 4.89EE190 pKa = 3.99DD191 pKa = 4.47DD192 pKa = 3.85EE193 pKa = 5.88VIFVSAKK200 pKa = 8.33PGGRR204 pKa = 11.84KK205 pKa = 9.11RR206 pKa = 11.84SAATPCEE213 pKa = 3.72PDD215 pKa = 3.5GASKK219 pKa = 10.52RR220 pKa = 11.84PCVPEE225 pKa = 4.13PEE227 pKa = 4.22QTEE230 pKa = 4.07PLDD233 pKa = 4.45LSLKK237 pKa = 9.67PRR239 pKa = 11.84PNN241 pKa = 3.21
MM1 pKa = 7.96KK2 pKa = 9.44YY3 pKa = 9.12TIVPAPRR10 pKa = 11.84NLHH13 pKa = 6.86DD14 pKa = 4.27YY15 pKa = 10.64VLEE18 pKa = 4.72LLEE21 pKa = 4.54EE22 pKa = 4.15WQPDD26 pKa = 4.08CLDD29 pKa = 3.45CEE31 pKa = 4.57YY32 pKa = 10.51PHH34 pKa = 7.12GSPSPPTLHH43 pKa = 7.48DD44 pKa = 4.17LFDD47 pKa = 4.14VEE49 pKa = 5.88LEE51 pKa = 4.34TSHH54 pKa = 7.55SPFVGLCDD62 pKa = 3.43SCAEE66 pKa = 4.27ADD68 pKa = 3.78TDD70 pKa = 4.26SSASTEE76 pKa = 4.03ADD78 pKa = 3.22SGFSPLSTPPVSPIPPHH95 pKa = 5.76PTSPASISDD104 pKa = 3.82DD105 pKa = 3.14MLLCLEE111 pKa = 4.63EE112 pKa = 4.67MPTFDD117 pKa = 6.17DD118 pKa = 3.65EE119 pKa = 5.53DD120 pKa = 4.02EE121 pKa = 4.35VRR123 pKa = 11.84SAATTFEE130 pKa = 3.86RR131 pKa = 11.84WEE133 pKa = 4.09NTFDD137 pKa = 3.56PHH139 pKa = 6.94VGPIFGCLRR148 pKa = 11.84CAFYY152 pKa = 10.82QEE154 pKa = 4.06QDD156 pKa = 3.54DD157 pKa = 4.13NALCGLCYY165 pKa = 10.43LKK167 pKa = 10.94ALAEE171 pKa = 4.38VPFALPVRR179 pKa = 11.84SEE181 pKa = 3.97PASAGAEE188 pKa = 4.06EE189 pKa = 4.89EE190 pKa = 3.99DD191 pKa = 4.47DD192 pKa = 3.85EE193 pKa = 5.88VIFVSAKK200 pKa = 8.33PGGRR204 pKa = 11.84KK205 pKa = 9.11RR206 pKa = 11.84SAATPCEE213 pKa = 3.72PDD215 pKa = 3.5GASKK219 pKa = 10.52RR220 pKa = 11.84PCVPEE225 pKa = 4.13PEE227 pKa = 4.22QTEE230 pKa = 4.07PLDD233 pKa = 4.45LSLKK237 pKa = 9.67PRR239 pKa = 11.84PNN241 pKa = 3.21
Molecular weight: 26.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6S6MFM5|A0A6S6MFM5_9ADEN E3 ORF A OS=Canine mastadenovirus A OX=10537 PE=4 SV=1
MM1 pKa = 7.77AGRR4 pKa = 11.84NVTLRR9 pKa = 11.84LRR11 pKa = 11.84VPVRR15 pKa = 11.84TKK17 pKa = 9.3ITGAGRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84GRR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84GIRR33 pKa = 11.84CGRR36 pKa = 11.84MRR38 pKa = 11.84GGFLPALIPLIAAAIGAVPGIASVALQAARR68 pKa = 11.84HH69 pKa = 4.96
MM1 pKa = 7.77AGRR4 pKa = 11.84NVTLRR9 pKa = 11.84LRR11 pKa = 11.84VPVRR15 pKa = 11.84TKK17 pKa = 9.3ITGAGRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84GRR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84GIRR33 pKa = 11.84CGRR36 pKa = 11.84MRR38 pKa = 11.84GGFLPALIPLIAAAIGAVPGIASVALQAARR68 pKa = 11.84HH69 pKa = 4.96
Molecular weight: 7.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10145 |
55 |
1150 |
327.3 |
36.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.777 ± 0.339 | 2.04 ± 0.309 |
4.406 ± 0.22 | 6.18 ± 0.45 |
4.268 ± 0.192 | 5.786 ± 0.36 |
2.326 ± 0.201 | 4.406 ± 0.213 |
4.593 ± 0.498 | 9.216 ± 0.345 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.642 ± 0.172 | 5.126 ± 0.431 |
6.516 ± 0.404 | 4.317 ± 0.211 |
5.954 ± 0.521 | 7.186 ± 0.234 |
6.082 ± 0.338 | 6.614 ± 0.292 |
1.242 ± 0.096 | 3.322 ± 0.329 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
