
Natranaerobius thermophilus (strain ATCC BAA-1301 / DSM 18059 / JW/NM-WN-LF)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Natranaerobiales; Natranaerobiaceae; Natranaerobius; Natranaerobius thermophilus
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
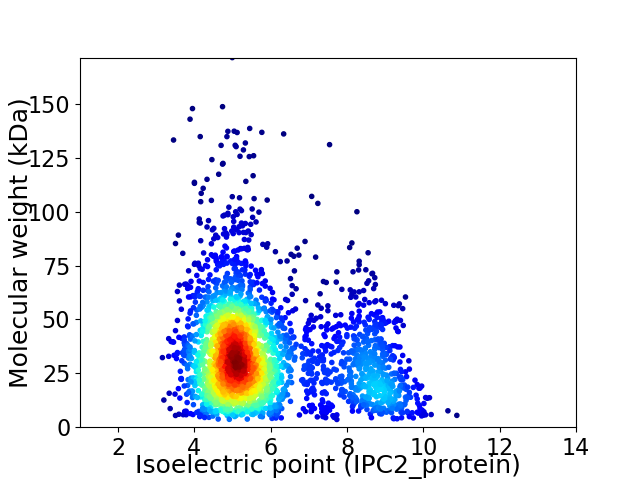
Virtual 2D-PAGE plot for 2847 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
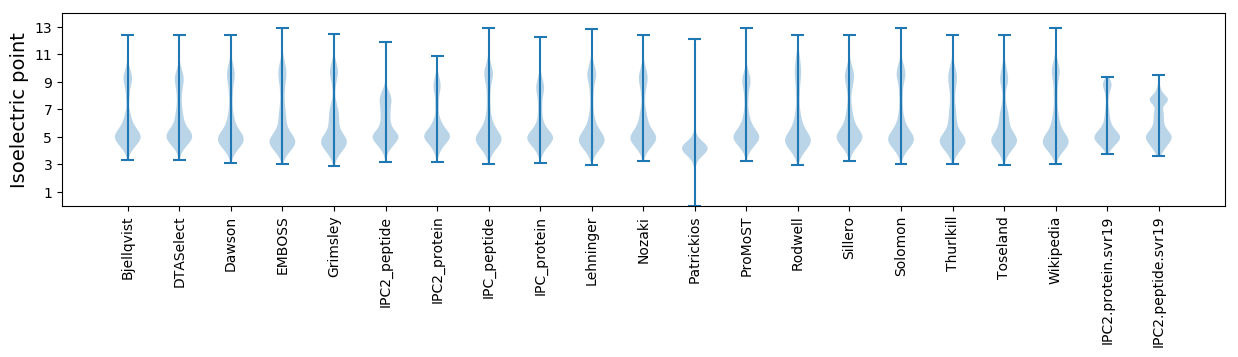
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B2A357|B2A357_NATTJ Flagellar protein FliL OS=Natranaerobius thermophilus (strain ATCC BAA-1301 / DSM 18059 / JW/NM-WN-LF) OX=457570 GN=Nther_1404 PE=3 SV=1
MM1 pKa = 7.81RR2 pKa = 11.84KK3 pKa = 9.49LLILLLVVFVASFGFMGCDD22 pKa = 2.96PADD25 pKa = 3.76PEE27 pKa = 4.4EE28 pKa = 4.2IAEE31 pKa = 4.25EE32 pKa = 4.29EE33 pKa = 4.17EE34 pKa = 4.38KK35 pKa = 11.04EE36 pKa = 4.19PEE38 pKa = 3.84EE39 pKa = 4.89DD40 pKa = 3.13VDD42 pKa = 4.05EE43 pKa = 4.76EE44 pKa = 4.53EE45 pKa = 5.08EE46 pKa = 4.12EE47 pKa = 4.29LEE49 pKa = 4.5QAITHH54 pKa = 6.98AIWSEE59 pKa = 3.53PDD61 pKa = 3.12GLFNYY66 pKa = 10.15CEE68 pKa = 4.09YY69 pKa = 10.96EE70 pKa = 3.94STYY73 pKa = 11.51DD74 pKa = 4.12LDD76 pKa = 4.34AFNQVFDD83 pKa = 4.4GMMEE87 pKa = 4.72ADD89 pKa = 3.44PHH91 pKa = 7.34ADD93 pKa = 4.11LEE95 pKa = 4.72LNPNLAEE102 pKa = 3.94EE103 pKa = 4.91HH104 pKa = 6.85EE105 pKa = 4.37ISDD108 pKa = 4.21DD109 pKa = 3.9GKK111 pKa = 8.59TFYY114 pKa = 11.2YY115 pKa = 10.65KK116 pKa = 10.69LRR118 pKa = 11.84DD119 pKa = 4.44DD120 pKa = 4.85IEE122 pKa = 4.4FHH124 pKa = 7.39DD125 pKa = 5.0GEE127 pKa = 4.86PLTAEE132 pKa = 3.91DD133 pKa = 4.33VKK135 pKa = 10.85FTFEE139 pKa = 4.15WMCHH143 pKa = 5.0EE144 pKa = 5.54DD145 pKa = 3.89YY146 pKa = 10.84IGPRR150 pKa = 11.84ASYY153 pKa = 8.11WQHH156 pKa = 7.44LEE158 pKa = 3.87GFDD161 pKa = 3.55EE162 pKa = 4.36YY163 pKa = 11.17RR164 pKa = 11.84AGEE167 pKa = 3.98ADD169 pKa = 3.56EE170 pKa = 4.7VEE172 pKa = 4.74GIEE175 pKa = 5.03IISDD179 pKa = 3.6HH180 pKa = 6.55EE181 pKa = 4.27IEE183 pKa = 4.25FHH185 pKa = 6.88FEE187 pKa = 3.87QVDD190 pKa = 3.67ASAVYY195 pKa = 10.16YY196 pKa = 10.6VSTWGISPKK205 pKa = 10.37HH206 pKa = 5.04VWEE209 pKa = 5.49DD210 pKa = 3.3IPVGEE215 pKa = 4.13RR216 pKa = 11.84RR217 pKa = 11.84EE218 pKa = 4.15APEE221 pKa = 3.59MTEE224 pKa = 4.42PIGTGPFEE232 pKa = 3.97FEE234 pKa = 4.52EE235 pKa = 4.51YY236 pKa = 11.08VEE238 pKa = 4.17GQYY241 pKa = 10.73VEE243 pKa = 5.52LVANEE248 pKa = 4.34DD249 pKa = 3.59YY250 pKa = 11.14HH251 pKa = 8.68RR252 pKa = 11.84GEE254 pKa = 4.68PEE256 pKa = 3.78LEE258 pKa = 4.55RR259 pKa = 11.84ITVEE263 pKa = 4.04VKK265 pKa = 10.77SPDD268 pKa = 3.4VVRR271 pKa = 11.84ADD273 pKa = 3.95LEE275 pKa = 4.3TGEE278 pKa = 4.23VDD280 pKa = 3.24IAEE283 pKa = 4.34VPPDD287 pKa = 3.34EE288 pKa = 5.64SEE290 pKa = 3.69WDD292 pKa = 3.57DD293 pKa = 4.32YY294 pKa = 11.34EE295 pKa = 4.8AHH297 pKa = 7.52DD298 pKa = 4.33EE299 pKa = 4.28LALEE303 pKa = 4.33YY304 pKa = 10.67YY305 pKa = 7.33PTNGYY310 pKa = 10.6QYY312 pKa = 10.33MGMNLRR318 pKa = 11.84DD319 pKa = 3.36EE320 pKa = 5.71SIFSDD325 pKa = 3.48HH326 pKa = 6.85AVRR329 pKa = 11.84EE330 pKa = 4.21AATYY334 pKa = 9.89AINRR338 pKa = 11.84EE339 pKa = 3.87GMVDD343 pKa = 4.46GILDD347 pKa = 3.76GLGKK351 pKa = 10.02VQNSHH356 pKa = 7.31FSPNQWAYY364 pKa = 10.99DD365 pKa = 4.13EE366 pKa = 5.96DD367 pKa = 5.88LDD369 pKa = 4.92TYY371 pKa = 11.15PHH373 pKa = 7.17DD374 pKa = 4.3PDD376 pKa = 3.43KK377 pKa = 11.64AEE379 pKa = 5.25EE380 pKa = 3.99ILEE383 pKa = 4.32DD384 pKa = 4.34AGYY387 pKa = 8.44TKK389 pKa = 10.9NDD391 pKa = 3.6DD392 pKa = 5.28GIWEE396 pKa = 4.24QNGEE400 pKa = 4.15PLEE403 pKa = 4.12FTLLYY408 pKa = 8.0PTGDD412 pKa = 3.56EE413 pKa = 4.17PRR415 pKa = 11.84EE416 pKa = 3.74QAALIIEE423 pKa = 4.12QDD425 pKa = 3.61LQGIGIDD432 pKa = 3.24ITLEE436 pKa = 3.93SLEE439 pKa = 4.3FATLSDD445 pKa = 3.55RR446 pKa = 11.84VFDD449 pKa = 5.33EE450 pKa = 4.35IDD452 pKa = 3.08FDD454 pKa = 4.68AYY456 pKa = 11.44LMGWSLGADD465 pKa = 3.81PDD467 pKa = 3.94PTGIWGPDD475 pKa = 2.91EE476 pKa = 4.44RR477 pKa = 11.84FNAVGFVHH485 pKa = 6.94PEE487 pKa = 3.55SDD489 pKa = 3.1EE490 pKa = 4.8LMEE493 pKa = 4.34QGLRR497 pKa = 11.84TTDD500 pKa = 3.09RR501 pKa = 11.84DD502 pKa = 3.35EE503 pKa = 4.6RR504 pKa = 11.84RR505 pKa = 11.84EE506 pKa = 4.4HH507 pKa = 5.55YY508 pKa = 10.85VEE510 pKa = 3.92WQRR513 pKa = 11.84LLQEE517 pKa = 3.79EE518 pKa = 4.5MPYY521 pKa = 10.61VFLYY525 pKa = 10.89ADD527 pKa = 3.77YY528 pKa = 11.15EE529 pKa = 4.81GYY531 pKa = 10.93AYY533 pKa = 11.19NNDD536 pKa = 2.95IQVFEE541 pKa = 4.6PNPWNIWYY549 pKa = 8.82DD550 pKa = 3.29VHH552 pKa = 6.54EE553 pKa = 5.02WYY555 pKa = 11.05LEE557 pKa = 3.89
MM1 pKa = 7.81RR2 pKa = 11.84KK3 pKa = 9.49LLILLLVVFVASFGFMGCDD22 pKa = 2.96PADD25 pKa = 3.76PEE27 pKa = 4.4EE28 pKa = 4.2IAEE31 pKa = 4.25EE32 pKa = 4.29EE33 pKa = 4.17EE34 pKa = 4.38KK35 pKa = 11.04EE36 pKa = 4.19PEE38 pKa = 3.84EE39 pKa = 4.89DD40 pKa = 3.13VDD42 pKa = 4.05EE43 pKa = 4.76EE44 pKa = 4.53EE45 pKa = 5.08EE46 pKa = 4.12EE47 pKa = 4.29LEE49 pKa = 4.5QAITHH54 pKa = 6.98AIWSEE59 pKa = 3.53PDD61 pKa = 3.12GLFNYY66 pKa = 10.15CEE68 pKa = 4.09YY69 pKa = 10.96EE70 pKa = 3.94STYY73 pKa = 11.51DD74 pKa = 4.12LDD76 pKa = 4.34AFNQVFDD83 pKa = 4.4GMMEE87 pKa = 4.72ADD89 pKa = 3.44PHH91 pKa = 7.34ADD93 pKa = 4.11LEE95 pKa = 4.72LNPNLAEE102 pKa = 3.94EE103 pKa = 4.91HH104 pKa = 6.85EE105 pKa = 4.37ISDD108 pKa = 4.21DD109 pKa = 3.9GKK111 pKa = 8.59TFYY114 pKa = 11.2YY115 pKa = 10.65KK116 pKa = 10.69LRR118 pKa = 11.84DD119 pKa = 4.44DD120 pKa = 4.85IEE122 pKa = 4.4FHH124 pKa = 7.39DD125 pKa = 5.0GEE127 pKa = 4.86PLTAEE132 pKa = 3.91DD133 pKa = 4.33VKK135 pKa = 10.85FTFEE139 pKa = 4.15WMCHH143 pKa = 5.0EE144 pKa = 5.54DD145 pKa = 3.89YY146 pKa = 10.84IGPRR150 pKa = 11.84ASYY153 pKa = 8.11WQHH156 pKa = 7.44LEE158 pKa = 3.87GFDD161 pKa = 3.55EE162 pKa = 4.36YY163 pKa = 11.17RR164 pKa = 11.84AGEE167 pKa = 3.98ADD169 pKa = 3.56EE170 pKa = 4.7VEE172 pKa = 4.74GIEE175 pKa = 5.03IISDD179 pKa = 3.6HH180 pKa = 6.55EE181 pKa = 4.27IEE183 pKa = 4.25FHH185 pKa = 6.88FEE187 pKa = 3.87QVDD190 pKa = 3.67ASAVYY195 pKa = 10.16YY196 pKa = 10.6VSTWGISPKK205 pKa = 10.37HH206 pKa = 5.04VWEE209 pKa = 5.49DD210 pKa = 3.3IPVGEE215 pKa = 4.13RR216 pKa = 11.84RR217 pKa = 11.84EE218 pKa = 4.15APEE221 pKa = 3.59MTEE224 pKa = 4.42PIGTGPFEE232 pKa = 3.97FEE234 pKa = 4.52EE235 pKa = 4.51YY236 pKa = 11.08VEE238 pKa = 4.17GQYY241 pKa = 10.73VEE243 pKa = 5.52LVANEE248 pKa = 4.34DD249 pKa = 3.59YY250 pKa = 11.14HH251 pKa = 8.68RR252 pKa = 11.84GEE254 pKa = 4.68PEE256 pKa = 3.78LEE258 pKa = 4.55RR259 pKa = 11.84ITVEE263 pKa = 4.04VKK265 pKa = 10.77SPDD268 pKa = 3.4VVRR271 pKa = 11.84ADD273 pKa = 3.95LEE275 pKa = 4.3TGEE278 pKa = 4.23VDD280 pKa = 3.24IAEE283 pKa = 4.34VPPDD287 pKa = 3.34EE288 pKa = 5.64SEE290 pKa = 3.69WDD292 pKa = 3.57DD293 pKa = 4.32YY294 pKa = 11.34EE295 pKa = 4.8AHH297 pKa = 7.52DD298 pKa = 4.33EE299 pKa = 4.28LALEE303 pKa = 4.33YY304 pKa = 10.67YY305 pKa = 7.33PTNGYY310 pKa = 10.6QYY312 pKa = 10.33MGMNLRR318 pKa = 11.84DD319 pKa = 3.36EE320 pKa = 5.71SIFSDD325 pKa = 3.48HH326 pKa = 6.85AVRR329 pKa = 11.84EE330 pKa = 4.21AATYY334 pKa = 9.89AINRR338 pKa = 11.84EE339 pKa = 3.87GMVDD343 pKa = 4.46GILDD347 pKa = 3.76GLGKK351 pKa = 10.02VQNSHH356 pKa = 7.31FSPNQWAYY364 pKa = 10.99DD365 pKa = 4.13EE366 pKa = 5.96DD367 pKa = 5.88LDD369 pKa = 4.92TYY371 pKa = 11.15PHH373 pKa = 7.17DD374 pKa = 4.3PDD376 pKa = 3.43KK377 pKa = 11.64AEE379 pKa = 5.25EE380 pKa = 3.99ILEE383 pKa = 4.32DD384 pKa = 4.34AGYY387 pKa = 8.44TKK389 pKa = 10.9NDD391 pKa = 3.6DD392 pKa = 5.28GIWEE396 pKa = 4.24QNGEE400 pKa = 4.15PLEE403 pKa = 4.12FTLLYY408 pKa = 8.0PTGDD412 pKa = 3.56EE413 pKa = 4.17PRR415 pKa = 11.84EE416 pKa = 3.74QAALIIEE423 pKa = 4.12QDD425 pKa = 3.61LQGIGIDD432 pKa = 3.24ITLEE436 pKa = 3.93SLEE439 pKa = 4.3FATLSDD445 pKa = 3.55RR446 pKa = 11.84VFDD449 pKa = 5.33EE450 pKa = 4.35IDD452 pKa = 3.08FDD454 pKa = 4.68AYY456 pKa = 11.44LMGWSLGADD465 pKa = 3.81PDD467 pKa = 3.94PTGIWGPDD475 pKa = 2.91EE476 pKa = 4.44RR477 pKa = 11.84FNAVGFVHH485 pKa = 6.94PEE487 pKa = 3.55SDD489 pKa = 3.1EE490 pKa = 4.8LMEE493 pKa = 4.34QGLRR497 pKa = 11.84TTDD500 pKa = 3.09RR501 pKa = 11.84DD502 pKa = 3.35EE503 pKa = 4.6RR504 pKa = 11.84RR505 pKa = 11.84EE506 pKa = 4.4HH507 pKa = 5.55YY508 pKa = 10.85VEE510 pKa = 3.92WQRR513 pKa = 11.84LLQEE517 pKa = 3.79EE518 pKa = 4.5MPYY521 pKa = 10.61VFLYY525 pKa = 10.89ADD527 pKa = 3.77YY528 pKa = 11.15EE529 pKa = 4.81GYY531 pKa = 10.93AYY533 pKa = 11.19NNDD536 pKa = 2.95IQVFEE541 pKa = 4.6PNPWNIWYY549 pKa = 8.82DD550 pKa = 3.29VHH552 pKa = 6.54EE553 pKa = 5.02WYY555 pKa = 11.05LEE557 pKa = 3.89
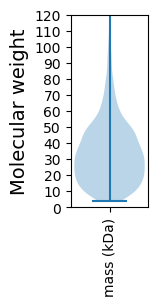
Molecular weight: 64.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2A498|B2A498_NATTJ Putative rRNA methylase OS=Natranaerobius thermophilus (strain ATCC BAA-1301 / DSM 18059 / JW/NM-WN-LF) OX=457570 GN=Nther_0153 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.0KK9 pKa = 7.87RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.39KK14 pKa = 8.49EE15 pKa = 3.5HH16 pKa = 6.11GFRR19 pKa = 11.84KK20 pKa = 9.77RR21 pKa = 11.84MKK23 pKa = 9.23TKK25 pKa = 10.17AGRR28 pKa = 11.84NILRR32 pKa = 11.84NRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.18GRR39 pKa = 11.84KK40 pKa = 6.43TLSAA44 pKa = 4.15
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.0KK9 pKa = 7.87RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.39KK14 pKa = 8.49EE15 pKa = 3.5HH16 pKa = 6.11GFRR19 pKa = 11.84KK20 pKa = 9.77RR21 pKa = 11.84MKK23 pKa = 9.23TKK25 pKa = 10.17AGRR28 pKa = 11.84NILRR32 pKa = 11.84NRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.18GRR39 pKa = 11.84KK40 pKa = 6.43TLSAA44 pKa = 4.15
Molecular weight: 5.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
893166 |
31 |
1474 |
313.7 |
35.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.063 ± 0.048 | 0.884 ± 0.019 |
5.848 ± 0.042 | 8.555 ± 0.068 |
4.126 ± 0.037 | 6.95 ± 0.047 |
1.837 ± 0.018 | 8.064 ± 0.049 |
6.829 ± 0.057 | 9.808 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.472 ± 0.021 | 4.969 ± 0.035 |
3.642 ± 0.022 | 3.827 ± 0.036 |
3.953 ± 0.03 | 6.028 ± 0.033 |
5.086 ± 0.028 | 6.671 ± 0.036 |
0.817 ± 0.016 | 3.573 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
