
Trichomonas vaginalis virus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Trichomonasvirus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
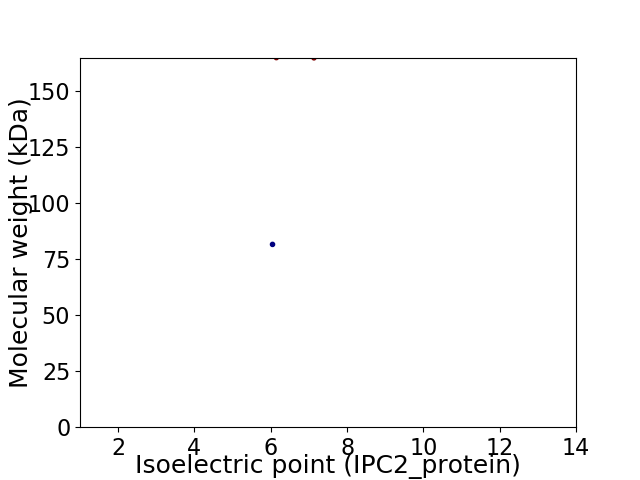
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F5B2U7|F5B2U7_9VIRU Capsid protein OS=Trichomonas vaginalis virus 4 OX=1008292 GN=cap PE=4 SV=1
MM1 pKa = 7.23SAIAATISSANLNDD15 pKa = 3.99LSRR18 pKa = 11.84LAGAQPKK25 pKa = 9.62EE26 pKa = 4.3GVPAPVLQQNIATPKK41 pKa = 9.49TGPPDD46 pKa = 4.05PGEE49 pKa = 4.23GTRR52 pKa = 11.84KK53 pKa = 8.1QTTDD57 pKa = 3.27SPHH60 pKa = 6.15SANPSTKK67 pKa = 9.08EE68 pKa = 3.65HH69 pKa = 6.28TPAPTIQPDD78 pKa = 4.02TPTPITDD85 pKa = 3.5HH86 pKa = 6.94SDD88 pKa = 3.4SEE90 pKa = 4.45EE91 pKa = 4.29GPRR94 pKa = 11.84THH96 pKa = 6.45STDD99 pKa = 3.35FQTLYY104 pKa = 10.64EE105 pKa = 4.26YY106 pKa = 10.32FYY108 pKa = 10.42SYY110 pKa = 10.38PVPASQTRR118 pKa = 11.84TGGAIARR125 pKa = 11.84AGPVNDD131 pKa = 3.51NNEE134 pKa = 4.31VVSFTTEE141 pKa = 3.68TALVTSLTPKK151 pKa = 10.48HH152 pKa = 6.32IDD154 pKa = 3.41ANIQPLQISIADD166 pKa = 3.5DD167 pKa = 3.68CVNYY171 pKa = 10.14SCQYY175 pKa = 10.63SGQTCPIFDD184 pKa = 4.37GSQHH188 pKa = 5.14VQSATALASSMKK200 pKa = 10.04ARR202 pKa = 11.84LMCEE206 pKa = 4.07VTQSLSARR214 pKa = 11.84PVQQPQLIAYY224 pKa = 9.26LYY226 pKa = 10.24GALLAFGDD234 pKa = 4.55RR235 pKa = 11.84LNIHH239 pKa = 6.43YY240 pKa = 10.2GNKK243 pKa = 8.68VNLWNALLGHH253 pKa = 6.41NLQRR257 pKa = 11.84GTPINGDD264 pKa = 3.45NFNHH268 pKa = 6.75HH269 pKa = 6.74LLIDD273 pKa = 4.43GPLAPPILPAAGLGPFPSTTLGPNTTVTFKK303 pKa = 10.9ARR305 pKa = 11.84ASIFVRR311 pKa = 11.84PQTYY315 pKa = 10.32DD316 pKa = 3.28YY317 pKa = 11.29ALVDD321 pKa = 3.37AAFWLIYY328 pKa = 10.99AMYY331 pKa = 10.7SRR333 pKa = 11.84MPVAFRR339 pKa = 11.84QAHH342 pKa = 5.4SLNIDD347 pKa = 4.35FFTVQPMAACVFPGHH362 pKa = 7.25DD363 pKa = 3.62GFTTPVIDD371 pKa = 3.47QALGVLEE378 pKa = 4.21SMLVEE383 pKa = 4.33MFNGDD388 pKa = 3.92RR389 pKa = 11.84EE390 pKa = 4.08IMYY393 pKa = 10.42YY394 pKa = 10.29YY395 pKa = 10.6AFKK398 pKa = 10.67GGQIFMRR405 pKa = 11.84PCSCYY410 pKa = 10.13QEE412 pKa = 4.78GGLIRR417 pKa = 11.84KK418 pKa = 9.32ASRR421 pKa = 11.84NVSLASFTGIYY432 pKa = 10.4SLIGYY437 pKa = 7.6CAPEE441 pKa = 4.22ARR443 pKa = 11.84PLHH446 pKa = 6.1AANHH450 pKa = 6.05PGIIAALFQYY460 pKa = 10.36VDD462 pKa = 3.64TMVLQAVLSYY472 pKa = 10.8SGPKK476 pKa = 9.52LVHH479 pKa = 6.62FGAAPEE485 pKa = 4.25FCSKK489 pKa = 10.9GSTPYY494 pKa = 11.26DD495 pKa = 4.12FIDD498 pKa = 4.31PDD500 pKa = 3.97NYY502 pKa = 8.86WGIRR506 pKa = 11.84AGVNAHH512 pKa = 7.03PIGYY516 pKa = 9.25YY517 pKa = 10.38YY518 pKa = 11.09LDD520 pKa = 2.88ILMRR524 pKa = 11.84PKK526 pKa = 10.12EE527 pKa = 4.06HH528 pKa = 6.58QLLDD532 pKa = 3.54EE533 pKa = 4.56TLSDD537 pKa = 3.31IYY539 pKa = 11.59GHH541 pKa = 6.03VGSLAMANIMASIASSGTEE560 pKa = 3.63VLNQKK565 pKa = 7.11MQKK568 pKa = 9.96SFVRR572 pKa = 11.84RR573 pKa = 11.84GNQVRR578 pKa = 11.84ALRR581 pKa = 11.84HH582 pKa = 4.08SHH584 pKa = 6.78AIINRR589 pKa = 11.84FHH591 pKa = 6.51EE592 pKa = 4.14PEE594 pKa = 3.61YY595 pKa = 10.65AYY597 pKa = 10.91RR598 pKa = 11.84LGILADD604 pKa = 4.75GIIPLAGTHH613 pKa = 6.14KK614 pKa = 10.66CDD616 pKa = 5.01IIDD619 pKa = 3.56EE620 pKa = 4.4ATRR623 pKa = 11.84LLQGEE628 pKa = 5.13DD629 pKa = 3.0IRR631 pKa = 11.84NLPGLRR637 pKa = 11.84CLRR640 pKa = 11.84GRR642 pKa = 11.84GLDD645 pKa = 3.62AIIGIRR651 pKa = 11.84PINKK655 pKa = 8.63KK656 pKa = 9.93RR657 pKa = 11.84RR658 pKa = 11.84AGFYY662 pKa = 9.33TLDD665 pKa = 3.73GNFHH669 pKa = 6.32VVTNQCTSDD678 pKa = 4.07VLQVWNDD685 pKa = 3.1HH686 pKa = 6.05GYY688 pKa = 9.43IARR691 pKa = 11.84PYY693 pKa = 8.38ACHH696 pKa = 6.11IVEE699 pKa = 4.77SINVEE704 pKa = 3.51IYY706 pKa = 10.48DD707 pKa = 4.23RR708 pKa = 11.84SNGAYY713 pKa = 10.12NGWIQALVSGFGVPEE728 pKa = 3.91RR729 pKa = 11.84CYY731 pKa = 9.94MGPRR735 pKa = 11.84LQVAGGAPSALL746 pKa = 3.66
MM1 pKa = 7.23SAIAATISSANLNDD15 pKa = 3.99LSRR18 pKa = 11.84LAGAQPKK25 pKa = 9.62EE26 pKa = 4.3GVPAPVLQQNIATPKK41 pKa = 9.49TGPPDD46 pKa = 4.05PGEE49 pKa = 4.23GTRR52 pKa = 11.84KK53 pKa = 8.1QTTDD57 pKa = 3.27SPHH60 pKa = 6.15SANPSTKK67 pKa = 9.08EE68 pKa = 3.65HH69 pKa = 6.28TPAPTIQPDD78 pKa = 4.02TPTPITDD85 pKa = 3.5HH86 pKa = 6.94SDD88 pKa = 3.4SEE90 pKa = 4.45EE91 pKa = 4.29GPRR94 pKa = 11.84THH96 pKa = 6.45STDD99 pKa = 3.35FQTLYY104 pKa = 10.64EE105 pKa = 4.26YY106 pKa = 10.32FYY108 pKa = 10.42SYY110 pKa = 10.38PVPASQTRR118 pKa = 11.84TGGAIARR125 pKa = 11.84AGPVNDD131 pKa = 3.51NNEE134 pKa = 4.31VVSFTTEE141 pKa = 3.68TALVTSLTPKK151 pKa = 10.48HH152 pKa = 6.32IDD154 pKa = 3.41ANIQPLQISIADD166 pKa = 3.5DD167 pKa = 3.68CVNYY171 pKa = 10.14SCQYY175 pKa = 10.63SGQTCPIFDD184 pKa = 4.37GSQHH188 pKa = 5.14VQSATALASSMKK200 pKa = 10.04ARR202 pKa = 11.84LMCEE206 pKa = 4.07VTQSLSARR214 pKa = 11.84PVQQPQLIAYY224 pKa = 9.26LYY226 pKa = 10.24GALLAFGDD234 pKa = 4.55RR235 pKa = 11.84LNIHH239 pKa = 6.43YY240 pKa = 10.2GNKK243 pKa = 8.68VNLWNALLGHH253 pKa = 6.41NLQRR257 pKa = 11.84GTPINGDD264 pKa = 3.45NFNHH268 pKa = 6.75HH269 pKa = 6.74LLIDD273 pKa = 4.43GPLAPPILPAAGLGPFPSTTLGPNTTVTFKK303 pKa = 10.9ARR305 pKa = 11.84ASIFVRR311 pKa = 11.84PQTYY315 pKa = 10.32DD316 pKa = 3.28YY317 pKa = 11.29ALVDD321 pKa = 3.37AAFWLIYY328 pKa = 10.99AMYY331 pKa = 10.7SRR333 pKa = 11.84MPVAFRR339 pKa = 11.84QAHH342 pKa = 5.4SLNIDD347 pKa = 4.35FFTVQPMAACVFPGHH362 pKa = 7.25DD363 pKa = 3.62GFTTPVIDD371 pKa = 3.47QALGVLEE378 pKa = 4.21SMLVEE383 pKa = 4.33MFNGDD388 pKa = 3.92RR389 pKa = 11.84EE390 pKa = 4.08IMYY393 pKa = 10.42YY394 pKa = 10.29YY395 pKa = 10.6AFKK398 pKa = 10.67GGQIFMRR405 pKa = 11.84PCSCYY410 pKa = 10.13QEE412 pKa = 4.78GGLIRR417 pKa = 11.84KK418 pKa = 9.32ASRR421 pKa = 11.84NVSLASFTGIYY432 pKa = 10.4SLIGYY437 pKa = 7.6CAPEE441 pKa = 4.22ARR443 pKa = 11.84PLHH446 pKa = 6.1AANHH450 pKa = 6.05PGIIAALFQYY460 pKa = 10.36VDD462 pKa = 3.64TMVLQAVLSYY472 pKa = 10.8SGPKK476 pKa = 9.52LVHH479 pKa = 6.62FGAAPEE485 pKa = 4.25FCSKK489 pKa = 10.9GSTPYY494 pKa = 11.26DD495 pKa = 4.12FIDD498 pKa = 4.31PDD500 pKa = 3.97NYY502 pKa = 8.86WGIRR506 pKa = 11.84AGVNAHH512 pKa = 7.03PIGYY516 pKa = 9.25YY517 pKa = 10.38YY518 pKa = 11.09LDD520 pKa = 2.88ILMRR524 pKa = 11.84PKK526 pKa = 10.12EE527 pKa = 4.06HH528 pKa = 6.58QLLDD532 pKa = 3.54EE533 pKa = 4.56TLSDD537 pKa = 3.31IYY539 pKa = 11.59GHH541 pKa = 6.03VGSLAMANIMASIASSGTEE560 pKa = 3.63VLNQKK565 pKa = 7.11MQKK568 pKa = 9.96SFVRR572 pKa = 11.84RR573 pKa = 11.84GNQVRR578 pKa = 11.84ALRR581 pKa = 11.84HH582 pKa = 4.08SHH584 pKa = 6.78AIINRR589 pKa = 11.84FHH591 pKa = 6.51EE592 pKa = 4.14PEE594 pKa = 3.61YY595 pKa = 10.65AYY597 pKa = 10.91RR598 pKa = 11.84LGILADD604 pKa = 4.75GIIPLAGTHH613 pKa = 6.14KK614 pKa = 10.66CDD616 pKa = 5.01IIDD619 pKa = 3.56EE620 pKa = 4.4ATRR623 pKa = 11.84LLQGEE628 pKa = 5.13DD629 pKa = 3.0IRR631 pKa = 11.84NLPGLRR637 pKa = 11.84CLRR640 pKa = 11.84GRR642 pKa = 11.84GLDD645 pKa = 3.62AIIGIRR651 pKa = 11.84PINKK655 pKa = 8.63KK656 pKa = 9.93RR657 pKa = 11.84RR658 pKa = 11.84AGFYY662 pKa = 9.33TLDD665 pKa = 3.73GNFHH669 pKa = 6.32VVTNQCTSDD678 pKa = 4.07VLQVWNDD685 pKa = 3.1HH686 pKa = 6.05GYY688 pKa = 9.43IARR691 pKa = 11.84PYY693 pKa = 8.38ACHH696 pKa = 6.11IVEE699 pKa = 4.77SINVEE704 pKa = 3.51IYY706 pKa = 10.48DD707 pKa = 4.23RR708 pKa = 11.84SNGAYY713 pKa = 10.12NGWIQALVSGFGVPEE728 pKa = 3.91RR729 pKa = 11.84CYY731 pKa = 9.94MGPRR735 pKa = 11.84LQVAGGAPSALL746 pKa = 3.66
Molecular weight: 81.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F5B2U7|F5B2U7_9VIRU Capsid protein OS=Trichomonas vaginalis virus 4 OX=1008292 GN=cap PE=4 SV=1
MM1 pKa = 7.23SAIAATISSANLNDD15 pKa = 3.99LSRR18 pKa = 11.84LAGAQPKK25 pKa = 9.62EE26 pKa = 4.3GVPAPVLQQNIATPKK41 pKa = 9.49TGPPDD46 pKa = 4.05PGEE49 pKa = 4.23GTRR52 pKa = 11.84KK53 pKa = 8.1QTTDD57 pKa = 3.27SPHH60 pKa = 6.15SANPSTKK67 pKa = 9.08EE68 pKa = 3.65HH69 pKa = 6.28TPAPTIQPDD78 pKa = 4.02TPTPITDD85 pKa = 3.5HH86 pKa = 6.94SDD88 pKa = 3.4SEE90 pKa = 4.45EE91 pKa = 4.29GPRR94 pKa = 11.84THH96 pKa = 6.45STDD99 pKa = 3.35FQTLYY104 pKa = 10.64EE105 pKa = 4.26YY106 pKa = 10.32FYY108 pKa = 10.42SYY110 pKa = 10.38PVPASQTRR118 pKa = 11.84TGGAIARR125 pKa = 11.84AGPVNDD131 pKa = 3.51NNEE134 pKa = 4.31VVSFTTEE141 pKa = 3.68TALVTSLTPKK151 pKa = 10.48HH152 pKa = 6.32IDD154 pKa = 3.41ANIQPLQISIADD166 pKa = 3.5DD167 pKa = 3.68CVNYY171 pKa = 10.14SCQYY175 pKa = 10.63SGQTCPIFDD184 pKa = 4.37GSQHH188 pKa = 5.14VQSATALASSMKK200 pKa = 10.04ARR202 pKa = 11.84LMCEE206 pKa = 4.07VTQSLSARR214 pKa = 11.84PVQQPQLIAYY224 pKa = 9.26LYY226 pKa = 10.24GALLAFGDD234 pKa = 4.55RR235 pKa = 11.84LNIHH239 pKa = 6.43YY240 pKa = 10.2GNKK243 pKa = 8.68VNLWNALLGHH253 pKa = 6.41NLQRR257 pKa = 11.84GTPINGDD264 pKa = 3.45NFNHH268 pKa = 6.75HH269 pKa = 6.74LLIDD273 pKa = 4.43GPLAPPILPAAGLGPFPSTTLGPNTTVTFKK303 pKa = 10.9ARR305 pKa = 11.84ASIFVRR311 pKa = 11.84PQTYY315 pKa = 10.32DD316 pKa = 3.28YY317 pKa = 11.29ALVDD321 pKa = 3.37AAFWLIYY328 pKa = 10.99AMYY331 pKa = 10.7SRR333 pKa = 11.84MPVAFRR339 pKa = 11.84QAHH342 pKa = 5.4SLNIDD347 pKa = 4.35FFTVQPMAACVFPGHH362 pKa = 7.25DD363 pKa = 3.62GFTTPVIDD371 pKa = 3.47QALGVLEE378 pKa = 4.21SMLVEE383 pKa = 4.33MFNGDD388 pKa = 3.92RR389 pKa = 11.84EE390 pKa = 4.08IMYY393 pKa = 10.42YY394 pKa = 10.29YY395 pKa = 10.6AFKK398 pKa = 10.67GGQIFMRR405 pKa = 11.84PCSCYY410 pKa = 10.13QEE412 pKa = 4.78GGLIRR417 pKa = 11.84KK418 pKa = 9.32ASRR421 pKa = 11.84NVSLASFTGIYY432 pKa = 10.4SLIGYY437 pKa = 7.6CAPEE441 pKa = 4.22ARR443 pKa = 11.84PLHH446 pKa = 6.1AANHH450 pKa = 6.05PGIIAALFQYY460 pKa = 10.36VDD462 pKa = 3.64TMVLQAVLSYY472 pKa = 10.8SGPKK476 pKa = 9.52LVHH479 pKa = 6.62FGAAPEE485 pKa = 4.25FCSKK489 pKa = 10.9GSTPYY494 pKa = 11.26DD495 pKa = 4.12FIDD498 pKa = 4.31PDD500 pKa = 3.97NYY502 pKa = 8.86WGIRR506 pKa = 11.84AGVNAHH512 pKa = 7.03PIGYY516 pKa = 9.25YY517 pKa = 10.38YY518 pKa = 11.09LDD520 pKa = 2.88ILMRR524 pKa = 11.84PKK526 pKa = 10.12EE527 pKa = 4.06HH528 pKa = 6.58QLLDD532 pKa = 3.54EE533 pKa = 4.56TLSDD537 pKa = 3.31IYY539 pKa = 11.59GHH541 pKa = 6.03VGSLAMANIMASIASSGTEE560 pKa = 3.63VLNQKK565 pKa = 7.11MQKK568 pKa = 9.96SFVRR572 pKa = 11.84RR573 pKa = 11.84GNQVRR578 pKa = 11.84ALRR581 pKa = 11.84HH582 pKa = 4.08SHH584 pKa = 6.78AIINRR589 pKa = 11.84FHH591 pKa = 6.51EE592 pKa = 4.14PEE594 pKa = 3.61YY595 pKa = 10.65AYY597 pKa = 10.91RR598 pKa = 11.84LGILADD604 pKa = 4.75GIIPLAGTHH613 pKa = 6.14KK614 pKa = 10.66CDD616 pKa = 5.01IIDD619 pKa = 3.56EE620 pKa = 4.4ATRR623 pKa = 11.84LLQGEE628 pKa = 5.13DD629 pKa = 3.0IRR631 pKa = 11.84NLPGLRR637 pKa = 11.84CLRR640 pKa = 11.84GRR642 pKa = 11.84GLDD645 pKa = 3.62AIIGIRR651 pKa = 11.84PINKK655 pKa = 8.63KK656 pKa = 9.93RR657 pKa = 11.84RR658 pKa = 11.84AGFYY662 pKa = 9.33TLDD665 pKa = 3.73GNFHH669 pKa = 6.32VVTNQCTSDD678 pKa = 4.07VLQVWNDD685 pKa = 3.1HH686 pKa = 6.05GYY688 pKa = 9.43IARR691 pKa = 11.84PYY693 pKa = 8.38ACHH696 pKa = 6.11IVEE699 pKa = 4.77SINVEE704 pKa = 3.51IYY706 pKa = 10.48DD707 pKa = 4.23RR708 pKa = 11.84SNGAYY713 pKa = 10.12NGWIQALVSGFGVPEE728 pKa = 3.95RR729 pKa = 11.84CYY731 pKa = 10.01MGPSSAGSRR740 pKa = 11.84RR741 pKa = 11.84RR742 pKa = 11.84PLCPLKK748 pKa = 10.9GSNCAVALHH757 pKa = 6.5VDD759 pKa = 3.92GQLTRR764 pKa = 11.84ASRR767 pKa = 11.84VPYY770 pKa = 10.26RR771 pKa = 11.84KK772 pKa = 8.41LTPSHH777 pKa = 6.57LNCSKK782 pKa = 10.67RR783 pKa = 11.84CARR786 pKa = 11.84QLAVIYY792 pKa = 9.8RR793 pKa = 11.84YY794 pKa = 7.97QTLSPQLTEE803 pKa = 4.65VSDD806 pKa = 3.69SDD808 pKa = 3.76YY809 pKa = 11.31LAFLRR814 pKa = 11.84WVLLPYY820 pKa = 10.07TGATNRR826 pKa = 11.84PHH828 pKa = 7.16PKK830 pKa = 9.26RR831 pKa = 11.84WPKK834 pKa = 9.9PFYY837 pKa = 9.72PAEE840 pKa = 4.06VSLKK844 pKa = 10.52FLDD847 pKa = 4.83KK848 pKa = 10.33KK849 pKa = 9.6TEE851 pKa = 3.97LQLFPLKK858 pKa = 10.33KK859 pKa = 10.44APQADD864 pKa = 3.61LKK866 pKa = 10.98VNCFARR872 pKa = 11.84NLLYY876 pKa = 10.64SSPLSDD882 pKa = 5.84RR883 pKa = 11.84ILKK886 pKa = 9.41QCIPVGTNNDD896 pKa = 4.0TVCGLVILLEE906 pKa = 4.29LLFEE910 pKa = 5.23AGVPLDD916 pKa = 4.25LLPTISVAIAKK927 pKa = 9.67NDD929 pKa = 3.75PFVKK933 pKa = 10.45ALSDD937 pKa = 3.81FNKK940 pKa = 8.05MTGATTSHH948 pKa = 7.01IANLLTEE955 pKa = 4.38CTTLLGRR962 pKa = 11.84GVTASAPNADD972 pKa = 4.23LYY974 pKa = 11.38HH975 pKa = 6.52RR976 pKa = 11.84VAPEE980 pKa = 4.07GNRR983 pKa = 11.84HH984 pKa = 5.02EE985 pKa = 5.29AKK987 pKa = 10.44ISDD990 pKa = 3.92DD991 pKa = 3.67VLRR994 pKa = 11.84SAIRR998 pKa = 11.84TIYY1001 pKa = 9.38KK1002 pKa = 9.77QEE1004 pKa = 4.1IKK1006 pKa = 10.46DD1007 pKa = 3.86CPKK1010 pKa = 10.34PGDD1013 pKa = 3.74FGLHH1017 pKa = 6.5LLTSPFWCKK1026 pKa = 10.31SGSHH1030 pKa = 6.32HH1031 pKa = 6.84HH1032 pKa = 6.34PQFPRR1037 pKa = 11.84YY1038 pKa = 8.93RR1039 pKa = 11.84NRR1041 pKa = 11.84LEE1043 pKa = 4.47FVMNTDD1049 pKa = 3.82PSAIMAVKK1057 pKa = 9.24PSVYY1061 pKa = 8.79ITQAQKK1067 pKa = 11.03LEE1069 pKa = 4.22HH1070 pKa = 6.43GKK1072 pKa = 7.86TRR1074 pKa = 11.84YY1075 pKa = 9.23IYY1077 pKa = 10.63NCDD1080 pKa = 3.39TVSYY1084 pKa = 10.46LYY1086 pKa = 10.47FDD1088 pKa = 4.73YY1089 pKa = 10.79ILNYY1093 pKa = 9.94VEE1095 pKa = 5.65SIWANSHH1102 pKa = 5.91VLLNPDD1108 pKa = 3.65ALNAEE1113 pKa = 4.35KK1114 pKa = 10.08FATLEE1119 pKa = 3.76YY1120 pKa = 10.73SEE1122 pKa = 5.08YY1123 pKa = 11.42CMIDD1127 pKa = 3.17YY1128 pKa = 10.72TDD1130 pKa = 4.84FNSQHH1135 pKa = 6.34TLTSMKK1141 pKa = 10.63AVFEE1145 pKa = 4.16VLKK1148 pKa = 10.54EE1149 pKa = 3.92FLPSEE1154 pKa = 4.3MFPVLDD1160 pKa = 3.49WCISSFDD1167 pKa = 3.52NMTIKK1172 pKa = 10.66DD1173 pKa = 3.95MKK1175 pKa = 9.7WRR1177 pKa = 11.84STLPSGHH1184 pKa = 7.18RR1185 pKa = 11.84ATTFINSVLNRR1196 pKa = 11.84AYY1198 pKa = 10.57LLPYY1202 pKa = 9.47IGTIVSYY1209 pKa = 10.79HH1210 pKa = 6.69CGDD1213 pKa = 4.02DD1214 pKa = 3.53VLLCGEE1220 pKa = 5.14HH1221 pKa = 7.66DD1222 pKa = 4.12YY1223 pKa = 11.44QHH1225 pKa = 7.39LITRR1229 pKa = 11.84LPYY1232 pKa = 9.87EE1233 pKa = 4.65LNPSKK1238 pKa = 10.86QSFGPHH1244 pKa = 6.07AEE1246 pKa = 3.77FLRR1249 pKa = 11.84LHH1251 pKa = 5.77RR1252 pKa = 11.84HH1253 pKa = 4.49GEE1255 pKa = 4.09KK1256 pKa = 10.66VIGYY1260 pKa = 5.44PTRR1263 pKa = 11.84AVSSLVSGNWLSTTSWNWQPSLLSITNQINAIICRR1298 pKa = 11.84SQLSISRR1305 pKa = 11.84IRR1307 pKa = 11.84SLAQEE1312 pKa = 3.97LRR1314 pKa = 11.84FRR1316 pKa = 11.84YY1317 pKa = 9.88CPLLDD1322 pKa = 4.18NYY1324 pKa = 9.91IDD1326 pKa = 4.1PATTSFVAAGCPSYY1340 pKa = 11.11QPTATMITPDD1350 pKa = 3.94VPHH1353 pKa = 7.19LDD1355 pKa = 3.48AEE1357 pKa = 4.52EE1358 pKa = 4.49VEE1360 pKa = 4.8FTQLHH1365 pKa = 5.14QLAEE1369 pKa = 4.35YY1370 pKa = 10.2AINTYY1375 pKa = 9.55PWLNSVEE1382 pKa = 4.5SVNQLVRR1389 pKa = 11.84SRR1391 pKa = 11.84MRR1393 pKa = 11.84KK1394 pKa = 7.58PAARR1398 pKa = 11.84DD1399 pKa = 2.9IHH1401 pKa = 7.36YY1402 pKa = 10.58SVLGPAIPLVSYY1414 pKa = 9.94HH1415 pKa = 5.75HH1416 pKa = 7.13HH1417 pKa = 7.37CDD1419 pKa = 3.41PMVVPLTRR1427 pKa = 11.84RR1428 pKa = 11.84YY1429 pKa = 10.02YY1430 pKa = 10.18PRR1432 pKa = 11.84DD1433 pKa = 3.28HH1434 pKa = 7.22LAPPITPQVLPPQPVFCDD1452 pKa = 3.54RR1453 pKa = 11.84DD1454 pKa = 3.58LSPIMALKK1462 pKa = 10.06IAPAGVAVKK1471 pKa = 8.66VTADD1475 pKa = 3.78RR1476 pKa = 11.84PIASAA1481 pKa = 3.89
MM1 pKa = 7.23SAIAATISSANLNDD15 pKa = 3.99LSRR18 pKa = 11.84LAGAQPKK25 pKa = 9.62EE26 pKa = 4.3GVPAPVLQQNIATPKK41 pKa = 9.49TGPPDD46 pKa = 4.05PGEE49 pKa = 4.23GTRR52 pKa = 11.84KK53 pKa = 8.1QTTDD57 pKa = 3.27SPHH60 pKa = 6.15SANPSTKK67 pKa = 9.08EE68 pKa = 3.65HH69 pKa = 6.28TPAPTIQPDD78 pKa = 4.02TPTPITDD85 pKa = 3.5HH86 pKa = 6.94SDD88 pKa = 3.4SEE90 pKa = 4.45EE91 pKa = 4.29GPRR94 pKa = 11.84THH96 pKa = 6.45STDD99 pKa = 3.35FQTLYY104 pKa = 10.64EE105 pKa = 4.26YY106 pKa = 10.32FYY108 pKa = 10.42SYY110 pKa = 10.38PVPASQTRR118 pKa = 11.84TGGAIARR125 pKa = 11.84AGPVNDD131 pKa = 3.51NNEE134 pKa = 4.31VVSFTTEE141 pKa = 3.68TALVTSLTPKK151 pKa = 10.48HH152 pKa = 6.32IDD154 pKa = 3.41ANIQPLQISIADD166 pKa = 3.5DD167 pKa = 3.68CVNYY171 pKa = 10.14SCQYY175 pKa = 10.63SGQTCPIFDD184 pKa = 4.37GSQHH188 pKa = 5.14VQSATALASSMKK200 pKa = 10.04ARR202 pKa = 11.84LMCEE206 pKa = 4.07VTQSLSARR214 pKa = 11.84PVQQPQLIAYY224 pKa = 9.26LYY226 pKa = 10.24GALLAFGDD234 pKa = 4.55RR235 pKa = 11.84LNIHH239 pKa = 6.43YY240 pKa = 10.2GNKK243 pKa = 8.68VNLWNALLGHH253 pKa = 6.41NLQRR257 pKa = 11.84GTPINGDD264 pKa = 3.45NFNHH268 pKa = 6.75HH269 pKa = 6.74LLIDD273 pKa = 4.43GPLAPPILPAAGLGPFPSTTLGPNTTVTFKK303 pKa = 10.9ARR305 pKa = 11.84ASIFVRR311 pKa = 11.84PQTYY315 pKa = 10.32DD316 pKa = 3.28YY317 pKa = 11.29ALVDD321 pKa = 3.37AAFWLIYY328 pKa = 10.99AMYY331 pKa = 10.7SRR333 pKa = 11.84MPVAFRR339 pKa = 11.84QAHH342 pKa = 5.4SLNIDD347 pKa = 4.35FFTVQPMAACVFPGHH362 pKa = 7.25DD363 pKa = 3.62GFTTPVIDD371 pKa = 3.47QALGVLEE378 pKa = 4.21SMLVEE383 pKa = 4.33MFNGDD388 pKa = 3.92RR389 pKa = 11.84EE390 pKa = 4.08IMYY393 pKa = 10.42YY394 pKa = 10.29YY395 pKa = 10.6AFKK398 pKa = 10.67GGQIFMRR405 pKa = 11.84PCSCYY410 pKa = 10.13QEE412 pKa = 4.78GGLIRR417 pKa = 11.84KK418 pKa = 9.32ASRR421 pKa = 11.84NVSLASFTGIYY432 pKa = 10.4SLIGYY437 pKa = 7.6CAPEE441 pKa = 4.22ARR443 pKa = 11.84PLHH446 pKa = 6.1AANHH450 pKa = 6.05PGIIAALFQYY460 pKa = 10.36VDD462 pKa = 3.64TMVLQAVLSYY472 pKa = 10.8SGPKK476 pKa = 9.52LVHH479 pKa = 6.62FGAAPEE485 pKa = 4.25FCSKK489 pKa = 10.9GSTPYY494 pKa = 11.26DD495 pKa = 4.12FIDD498 pKa = 4.31PDD500 pKa = 3.97NYY502 pKa = 8.86WGIRR506 pKa = 11.84AGVNAHH512 pKa = 7.03PIGYY516 pKa = 9.25YY517 pKa = 10.38YY518 pKa = 11.09LDD520 pKa = 2.88ILMRR524 pKa = 11.84PKK526 pKa = 10.12EE527 pKa = 4.06HH528 pKa = 6.58QLLDD532 pKa = 3.54EE533 pKa = 4.56TLSDD537 pKa = 3.31IYY539 pKa = 11.59GHH541 pKa = 6.03VGSLAMANIMASIASSGTEE560 pKa = 3.63VLNQKK565 pKa = 7.11MQKK568 pKa = 9.96SFVRR572 pKa = 11.84RR573 pKa = 11.84GNQVRR578 pKa = 11.84ALRR581 pKa = 11.84HH582 pKa = 4.08SHH584 pKa = 6.78AIINRR589 pKa = 11.84FHH591 pKa = 6.51EE592 pKa = 4.14PEE594 pKa = 3.61YY595 pKa = 10.65AYY597 pKa = 10.91RR598 pKa = 11.84LGILADD604 pKa = 4.75GIIPLAGTHH613 pKa = 6.14KK614 pKa = 10.66CDD616 pKa = 5.01IIDD619 pKa = 3.56EE620 pKa = 4.4ATRR623 pKa = 11.84LLQGEE628 pKa = 5.13DD629 pKa = 3.0IRR631 pKa = 11.84NLPGLRR637 pKa = 11.84CLRR640 pKa = 11.84GRR642 pKa = 11.84GLDD645 pKa = 3.62AIIGIRR651 pKa = 11.84PINKK655 pKa = 8.63KK656 pKa = 9.93RR657 pKa = 11.84RR658 pKa = 11.84AGFYY662 pKa = 9.33TLDD665 pKa = 3.73GNFHH669 pKa = 6.32VVTNQCTSDD678 pKa = 4.07VLQVWNDD685 pKa = 3.1HH686 pKa = 6.05GYY688 pKa = 9.43IARR691 pKa = 11.84PYY693 pKa = 8.38ACHH696 pKa = 6.11IVEE699 pKa = 4.77SINVEE704 pKa = 3.51IYY706 pKa = 10.48DD707 pKa = 4.23RR708 pKa = 11.84SNGAYY713 pKa = 10.12NGWIQALVSGFGVPEE728 pKa = 3.95RR729 pKa = 11.84CYY731 pKa = 10.01MGPSSAGSRR740 pKa = 11.84RR741 pKa = 11.84RR742 pKa = 11.84PLCPLKK748 pKa = 10.9GSNCAVALHH757 pKa = 6.5VDD759 pKa = 3.92GQLTRR764 pKa = 11.84ASRR767 pKa = 11.84VPYY770 pKa = 10.26RR771 pKa = 11.84KK772 pKa = 8.41LTPSHH777 pKa = 6.57LNCSKK782 pKa = 10.67RR783 pKa = 11.84CARR786 pKa = 11.84QLAVIYY792 pKa = 9.8RR793 pKa = 11.84YY794 pKa = 7.97QTLSPQLTEE803 pKa = 4.65VSDD806 pKa = 3.69SDD808 pKa = 3.76YY809 pKa = 11.31LAFLRR814 pKa = 11.84WVLLPYY820 pKa = 10.07TGATNRR826 pKa = 11.84PHH828 pKa = 7.16PKK830 pKa = 9.26RR831 pKa = 11.84WPKK834 pKa = 9.9PFYY837 pKa = 9.72PAEE840 pKa = 4.06VSLKK844 pKa = 10.52FLDD847 pKa = 4.83KK848 pKa = 10.33KK849 pKa = 9.6TEE851 pKa = 3.97LQLFPLKK858 pKa = 10.33KK859 pKa = 10.44APQADD864 pKa = 3.61LKK866 pKa = 10.98VNCFARR872 pKa = 11.84NLLYY876 pKa = 10.64SSPLSDD882 pKa = 5.84RR883 pKa = 11.84ILKK886 pKa = 9.41QCIPVGTNNDD896 pKa = 4.0TVCGLVILLEE906 pKa = 4.29LLFEE910 pKa = 5.23AGVPLDD916 pKa = 4.25LLPTISVAIAKK927 pKa = 9.67NDD929 pKa = 3.75PFVKK933 pKa = 10.45ALSDD937 pKa = 3.81FNKK940 pKa = 8.05MTGATTSHH948 pKa = 7.01IANLLTEE955 pKa = 4.38CTTLLGRR962 pKa = 11.84GVTASAPNADD972 pKa = 4.23LYY974 pKa = 11.38HH975 pKa = 6.52RR976 pKa = 11.84VAPEE980 pKa = 4.07GNRR983 pKa = 11.84HH984 pKa = 5.02EE985 pKa = 5.29AKK987 pKa = 10.44ISDD990 pKa = 3.92DD991 pKa = 3.67VLRR994 pKa = 11.84SAIRR998 pKa = 11.84TIYY1001 pKa = 9.38KK1002 pKa = 9.77QEE1004 pKa = 4.1IKK1006 pKa = 10.46DD1007 pKa = 3.86CPKK1010 pKa = 10.34PGDD1013 pKa = 3.74FGLHH1017 pKa = 6.5LLTSPFWCKK1026 pKa = 10.31SGSHH1030 pKa = 6.32HH1031 pKa = 6.84HH1032 pKa = 6.34PQFPRR1037 pKa = 11.84YY1038 pKa = 8.93RR1039 pKa = 11.84NRR1041 pKa = 11.84LEE1043 pKa = 4.47FVMNTDD1049 pKa = 3.82PSAIMAVKK1057 pKa = 9.24PSVYY1061 pKa = 8.79ITQAQKK1067 pKa = 11.03LEE1069 pKa = 4.22HH1070 pKa = 6.43GKK1072 pKa = 7.86TRR1074 pKa = 11.84YY1075 pKa = 9.23IYY1077 pKa = 10.63NCDD1080 pKa = 3.39TVSYY1084 pKa = 10.46LYY1086 pKa = 10.47FDD1088 pKa = 4.73YY1089 pKa = 10.79ILNYY1093 pKa = 9.94VEE1095 pKa = 5.65SIWANSHH1102 pKa = 5.91VLLNPDD1108 pKa = 3.65ALNAEE1113 pKa = 4.35KK1114 pKa = 10.08FATLEE1119 pKa = 3.76YY1120 pKa = 10.73SEE1122 pKa = 5.08YY1123 pKa = 11.42CMIDD1127 pKa = 3.17YY1128 pKa = 10.72TDD1130 pKa = 4.84FNSQHH1135 pKa = 6.34TLTSMKK1141 pKa = 10.63AVFEE1145 pKa = 4.16VLKK1148 pKa = 10.54EE1149 pKa = 3.92FLPSEE1154 pKa = 4.3MFPVLDD1160 pKa = 3.49WCISSFDD1167 pKa = 3.52NMTIKK1172 pKa = 10.66DD1173 pKa = 3.95MKK1175 pKa = 9.7WRR1177 pKa = 11.84STLPSGHH1184 pKa = 7.18RR1185 pKa = 11.84ATTFINSVLNRR1196 pKa = 11.84AYY1198 pKa = 10.57LLPYY1202 pKa = 9.47IGTIVSYY1209 pKa = 10.79HH1210 pKa = 6.69CGDD1213 pKa = 4.02DD1214 pKa = 3.53VLLCGEE1220 pKa = 5.14HH1221 pKa = 7.66DD1222 pKa = 4.12YY1223 pKa = 11.44QHH1225 pKa = 7.39LITRR1229 pKa = 11.84LPYY1232 pKa = 9.87EE1233 pKa = 4.65LNPSKK1238 pKa = 10.86QSFGPHH1244 pKa = 6.07AEE1246 pKa = 3.77FLRR1249 pKa = 11.84LHH1251 pKa = 5.77RR1252 pKa = 11.84HH1253 pKa = 4.49GEE1255 pKa = 4.09KK1256 pKa = 10.66VIGYY1260 pKa = 5.44PTRR1263 pKa = 11.84AVSSLVSGNWLSTTSWNWQPSLLSITNQINAIICRR1298 pKa = 11.84SQLSISRR1305 pKa = 11.84IRR1307 pKa = 11.84SLAQEE1312 pKa = 3.97LRR1314 pKa = 11.84FRR1316 pKa = 11.84YY1317 pKa = 9.88CPLLDD1322 pKa = 4.18NYY1324 pKa = 9.91IDD1326 pKa = 4.1PATTSFVAAGCPSYY1340 pKa = 11.11QPTATMITPDD1350 pKa = 3.94VPHH1353 pKa = 7.19LDD1355 pKa = 3.48AEE1357 pKa = 4.52EE1358 pKa = 4.49VEE1360 pKa = 4.8FTQLHH1365 pKa = 5.14QLAEE1369 pKa = 4.35YY1370 pKa = 10.2AINTYY1375 pKa = 9.55PWLNSVEE1382 pKa = 4.5SVNQLVRR1389 pKa = 11.84SRR1391 pKa = 11.84MRR1393 pKa = 11.84KK1394 pKa = 7.58PAARR1398 pKa = 11.84DD1399 pKa = 2.9IHH1401 pKa = 7.36YY1402 pKa = 10.58SVLGPAIPLVSYY1414 pKa = 9.94HH1415 pKa = 5.75HH1416 pKa = 7.13HH1417 pKa = 7.37CDD1419 pKa = 3.41PMVVPLTRR1427 pKa = 11.84RR1428 pKa = 11.84YY1429 pKa = 10.02YY1430 pKa = 10.18PRR1432 pKa = 11.84DD1433 pKa = 3.28HH1434 pKa = 7.22LAPPITPQVLPPQPVFCDD1452 pKa = 3.54RR1453 pKa = 11.84DD1454 pKa = 3.58LSPIMALKK1462 pKa = 10.06IAPAGVAVKK1471 pKa = 8.66VTADD1475 pKa = 3.78RR1476 pKa = 11.84PIASAA1481 pKa = 3.89
Molecular weight: 164.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2227 |
746 |
1481 |
1113.5 |
123.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.936 ± 0.464 | 2.155 ± 0.152 |
4.984 ± 0.013 | 3.502 ± 0.083 |
3.682 ± 0.039 | 6.466 ± 0.862 |
3.458 ± 0.058 | 6.331 ± 0.276 |
3.098 ± 0.375 | 9.295 ± 0.538 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.976 ± 0.092 | 4.67 ± 0.085 |
7.454 ± 0.118 | 4.176 ± 0.282 |
5.344 ± 0.136 | 7.05 ± 0.263 |
6.152 ± 0.065 | 5.658 ± 0.162 |
0.898 ± 0.124 | 4.715 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
