
Hubei tombus-like virus 27
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.09
Get precalculated fractions of proteins
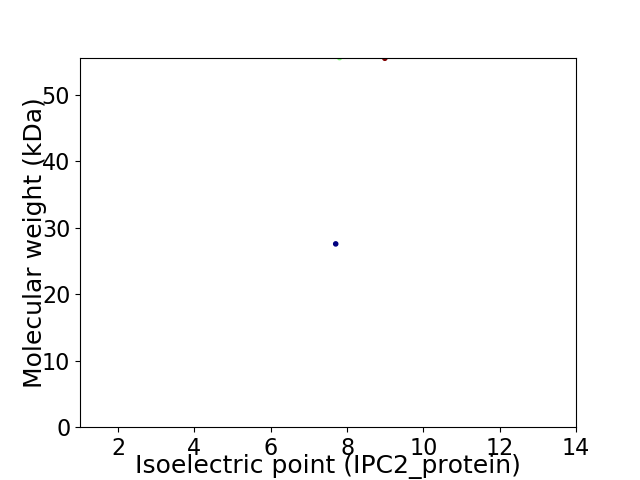
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG61|A0A1L3KG61_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 27 OX=1923274 PE=4 SV=1
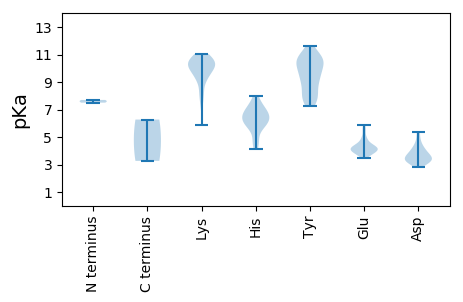
MM1 pKa = 7.5EE2 pKa = 5.56SIKK5 pKa = 10.98AFAQRR10 pKa = 11.84ISGWMMPKK18 pKa = 8.88KK19 pKa = 9.15TDD21 pKa = 3.78LQNQLDD27 pKa = 4.49TIKK30 pKa = 10.35YY31 pKa = 8.1IQMLLRR37 pKa = 11.84SKK39 pKa = 10.3PGQEE43 pKa = 4.18EE44 pKa = 3.76FHH46 pKa = 7.26DD47 pKa = 4.82AVDD50 pKa = 3.87NLNACKK56 pKa = 9.8TGPQIKK62 pKa = 8.21TVEE65 pKa = 4.41DD66 pKa = 4.04YY67 pKa = 11.64EE68 pKa = 4.8NDD70 pKa = 3.11ARR72 pKa = 11.84INAIRR77 pKa = 11.84SANQEE82 pKa = 3.58AALNHH87 pKa = 6.29LNYY90 pKa = 10.39KK91 pKa = 10.49SIGKK95 pKa = 9.75FIDD98 pKa = 3.35EE99 pKa = 5.85NYY101 pKa = 7.23TTWCAATLITYY112 pKa = 10.24LDD114 pKa = 3.66TVLITDD120 pKa = 3.31EE121 pKa = 4.46KK122 pKa = 10.51EE123 pKa = 3.93RR124 pKa = 11.84YY125 pKa = 9.75DD126 pKa = 4.26IISYY130 pKa = 8.71CIKK133 pKa = 10.21MNIAHH138 pKa = 6.41RR139 pKa = 11.84TANEE143 pKa = 3.73RR144 pKa = 11.84FIGEE148 pKa = 4.19NNFNDD153 pKa = 3.51IEE155 pKa = 4.15RR156 pKa = 11.84LNEE159 pKa = 3.81SSLTITRR166 pKa = 11.84DD167 pKa = 3.56KK168 pKa = 10.49PWWAFGFEE176 pKa = 4.22RR177 pKa = 11.84TRR179 pKa = 11.84GLTTKK184 pKa = 10.97ADD186 pKa = 3.91LNKK189 pKa = 10.11NRR191 pKa = 11.84ALVWCFRR198 pKa = 11.84PSTTSIIISTLGAIVIIDD216 pKa = 3.88KK217 pKa = 10.85LRR219 pKa = 11.84TIQQFKK225 pKa = 8.3TLRR228 pKa = 11.84NTITTLQAVNHH239 pKa = 6.27
MM1 pKa = 7.5EE2 pKa = 5.56SIKK5 pKa = 10.98AFAQRR10 pKa = 11.84ISGWMMPKK18 pKa = 8.88KK19 pKa = 9.15TDD21 pKa = 3.78LQNQLDD27 pKa = 4.49TIKK30 pKa = 10.35YY31 pKa = 8.1IQMLLRR37 pKa = 11.84SKK39 pKa = 10.3PGQEE43 pKa = 4.18EE44 pKa = 3.76FHH46 pKa = 7.26DD47 pKa = 4.82AVDD50 pKa = 3.87NLNACKK56 pKa = 9.8TGPQIKK62 pKa = 8.21TVEE65 pKa = 4.41DD66 pKa = 4.04YY67 pKa = 11.64EE68 pKa = 4.8NDD70 pKa = 3.11ARR72 pKa = 11.84INAIRR77 pKa = 11.84SANQEE82 pKa = 3.58AALNHH87 pKa = 6.29LNYY90 pKa = 10.39KK91 pKa = 10.49SIGKK95 pKa = 9.75FIDD98 pKa = 3.35EE99 pKa = 5.85NYY101 pKa = 7.23TTWCAATLITYY112 pKa = 10.24LDD114 pKa = 3.66TVLITDD120 pKa = 3.31EE121 pKa = 4.46KK122 pKa = 10.51EE123 pKa = 3.93RR124 pKa = 11.84YY125 pKa = 9.75DD126 pKa = 4.26IISYY130 pKa = 8.71CIKK133 pKa = 10.21MNIAHH138 pKa = 6.41RR139 pKa = 11.84TANEE143 pKa = 3.73RR144 pKa = 11.84FIGEE148 pKa = 4.19NNFNDD153 pKa = 3.51IEE155 pKa = 4.15RR156 pKa = 11.84LNEE159 pKa = 3.81SSLTITRR166 pKa = 11.84DD167 pKa = 3.56KK168 pKa = 10.49PWWAFGFEE176 pKa = 4.22RR177 pKa = 11.84TRR179 pKa = 11.84GLTTKK184 pKa = 10.97ADD186 pKa = 3.91LNKK189 pKa = 10.11NRR191 pKa = 11.84ALVWCFRR198 pKa = 11.84PSTTSIIISTLGAIVIIDD216 pKa = 3.88KK217 pKa = 10.85LRR219 pKa = 11.84TIQQFKK225 pKa = 8.3TLRR228 pKa = 11.84NTITTLQAVNHH239 pKa = 6.27
Molecular weight: 27.59 kDa
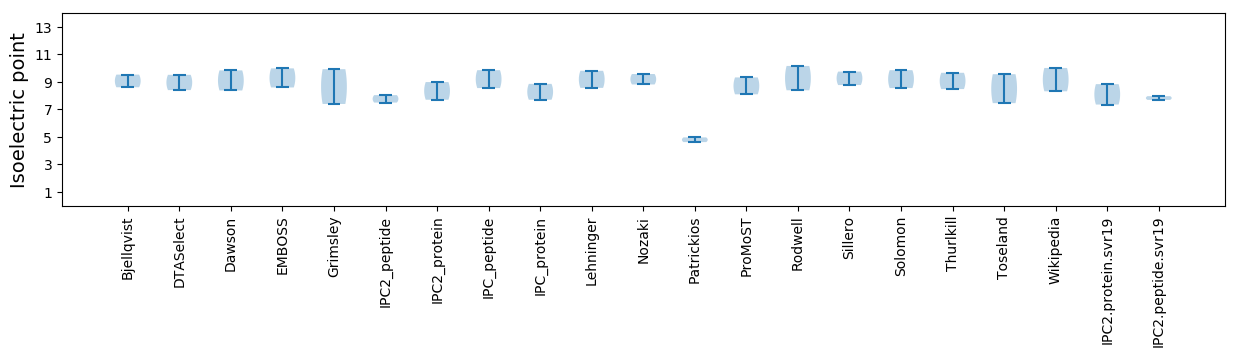
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG61|A0A1L3KG61_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 27 OX=1923274 PE=4 SV=1
MM1 pKa = 7.7VFQTVNNEE9 pKa = 3.83HH10 pKa = 6.41NNINFRR16 pKa = 11.84SDD18 pKa = 2.8RR19 pKa = 11.84YY20 pKa = 9.8NRR22 pKa = 11.84QIAHH26 pKa = 6.16YY27 pKa = 8.94PAIQDD32 pKa = 3.54PAQYY36 pKa = 10.49YY37 pKa = 7.76YY38 pKa = 10.71YY39 pKa = 9.64PSSRR43 pKa = 11.84EE44 pKa = 3.87SLIAAIKK51 pKa = 9.84NRR53 pKa = 11.84HH54 pKa = 4.66EE55 pKa = 4.0PEE57 pKa = 3.81RR58 pKa = 11.84TKK60 pKa = 10.77LYY62 pKa = 10.11QPKK65 pKa = 9.28IQKK68 pKa = 10.13NIYY71 pKa = 7.84KK72 pKa = 10.04HH73 pKa = 4.77IRR75 pKa = 11.84KK76 pKa = 9.22FIMKK80 pKa = 7.54VQPWTRR86 pKa = 11.84QQYY89 pKa = 9.12VDD91 pKa = 5.36AIIDD95 pKa = 3.54PGKK98 pKa = 10.07RR99 pKa = 11.84KK100 pKa = 9.97SYY102 pKa = 10.53QDD104 pKa = 3.85CLDD107 pKa = 3.32EE108 pKa = 4.23MEE110 pKa = 4.58RR111 pKa = 11.84TGKK114 pKa = 10.26VYY116 pKa = 10.71SHH118 pKa = 7.21IVPHH122 pKa = 5.76TKK124 pKa = 9.25IEE126 pKa = 4.07KK127 pKa = 9.35MKK129 pKa = 10.0SNKK132 pKa = 9.44YY133 pKa = 9.04KK134 pKa = 10.68APRR137 pKa = 11.84LIQARR142 pKa = 11.84HH143 pKa = 4.15MTHH146 pKa = 6.35NIEE149 pKa = 3.97VGRR152 pKa = 11.84YY153 pKa = 7.78LKK155 pKa = 10.4PLEE158 pKa = 4.17IAVKK162 pKa = 10.41SKK164 pKa = 10.64FINFTKK170 pKa = 10.64GNYY173 pKa = 9.63DD174 pKa = 3.18SVAARR179 pKa = 11.84IEE181 pKa = 3.99KK182 pKa = 10.03LASKK186 pKa = 9.99FKK188 pKa = 10.85YY189 pKa = 8.81FTEE192 pKa = 4.35SDD194 pKa = 3.45HH195 pKa = 5.68VTFDD199 pKa = 3.04AHH201 pKa = 5.82ITVEE205 pKa = 4.21HH206 pKa = 6.72LKK208 pKa = 8.33MTHH211 pKa = 6.38KK212 pKa = 10.68FYY214 pKa = 10.85LACFNHH220 pKa = 6.91DD221 pKa = 3.3RR222 pKa = 11.84TLQKK226 pKa = 10.68YY227 pKa = 9.31LKK229 pKa = 8.96KK230 pKa = 9.36TINNRR235 pKa = 11.84IRR237 pKa = 11.84TRR239 pKa = 11.84DD240 pKa = 3.36GTKK243 pKa = 7.21WTVRR247 pKa = 11.84GTRR250 pKa = 11.84MSGDD254 pKa = 3.13VDD256 pKa = 3.61TSFGNSLINYY266 pKa = 7.82AIIMQVLEE274 pKa = 3.82NLQLKK279 pKa = 10.19GDD281 pKa = 4.67AIVNGDD287 pKa = 3.79DD288 pKa = 5.22SIIFTNQKK296 pKa = 9.67VDD298 pKa = 3.25NKK300 pKa = 9.89LAKK303 pKa = 10.27IMFDD307 pKa = 3.19IYY309 pKa = 11.14NQEE312 pKa = 4.1TEE314 pKa = 4.12IKK316 pKa = 9.42EE317 pKa = 4.33SEE319 pKa = 4.12NSIHH323 pKa = 6.86RR324 pKa = 11.84VEE326 pKa = 4.88FCRR329 pKa = 11.84TRR331 pKa = 11.84LVYY334 pKa = 9.55PASGKK339 pKa = 7.71PTMMMDD345 pKa = 4.2PKK347 pKa = 10.22RR348 pKa = 11.84LNSIYY353 pKa = 10.92GMTYY357 pKa = 10.61KK358 pKa = 10.88LIPPEE363 pKa = 4.28EE364 pKa = 4.06YY365 pKa = 10.78ANYY368 pKa = 10.26LINIKK373 pKa = 9.93KK374 pKa = 10.53ANEE377 pKa = 4.42SINKK381 pKa = 5.87NTPIGHH387 pKa = 6.05YY388 pKa = 8.15WHH390 pKa = 6.94TDD392 pKa = 3.05EE393 pKa = 5.52EE394 pKa = 4.86VKK396 pKa = 10.38HH397 pKa = 6.68DD398 pKa = 3.53KK399 pKa = 11.05HH400 pKa = 8.0IDD402 pKa = 3.45YY403 pKa = 10.66SLKK406 pKa = 10.47RR407 pKa = 11.84VITRR411 pKa = 11.84EE412 pKa = 4.19SVNQTSTTEE421 pKa = 3.46ITITMFIAYY430 pKa = 7.67PNLLEE435 pKa = 5.12DD436 pKa = 4.18LNKK439 pKa = 8.16TTEE442 pKa = 4.56LIKK445 pKa = 10.91RR446 pKa = 11.84NLPRR450 pKa = 11.84KK451 pKa = 9.66NSPSWFIDD459 pKa = 3.1HH460 pKa = 7.56DD461 pKa = 4.27NKK463 pKa = 10.74KK464 pKa = 9.95LHH466 pKa = 5.87QLNDD470 pKa = 3.26
MM1 pKa = 7.7VFQTVNNEE9 pKa = 3.83HH10 pKa = 6.41NNINFRR16 pKa = 11.84SDD18 pKa = 2.8RR19 pKa = 11.84YY20 pKa = 9.8NRR22 pKa = 11.84QIAHH26 pKa = 6.16YY27 pKa = 8.94PAIQDD32 pKa = 3.54PAQYY36 pKa = 10.49YY37 pKa = 7.76YY38 pKa = 10.71YY39 pKa = 9.64PSSRR43 pKa = 11.84EE44 pKa = 3.87SLIAAIKK51 pKa = 9.84NRR53 pKa = 11.84HH54 pKa = 4.66EE55 pKa = 4.0PEE57 pKa = 3.81RR58 pKa = 11.84TKK60 pKa = 10.77LYY62 pKa = 10.11QPKK65 pKa = 9.28IQKK68 pKa = 10.13NIYY71 pKa = 7.84KK72 pKa = 10.04HH73 pKa = 4.77IRR75 pKa = 11.84KK76 pKa = 9.22FIMKK80 pKa = 7.54VQPWTRR86 pKa = 11.84QQYY89 pKa = 9.12VDD91 pKa = 5.36AIIDD95 pKa = 3.54PGKK98 pKa = 10.07RR99 pKa = 11.84KK100 pKa = 9.97SYY102 pKa = 10.53QDD104 pKa = 3.85CLDD107 pKa = 3.32EE108 pKa = 4.23MEE110 pKa = 4.58RR111 pKa = 11.84TGKK114 pKa = 10.26VYY116 pKa = 10.71SHH118 pKa = 7.21IVPHH122 pKa = 5.76TKK124 pKa = 9.25IEE126 pKa = 4.07KK127 pKa = 9.35MKK129 pKa = 10.0SNKK132 pKa = 9.44YY133 pKa = 9.04KK134 pKa = 10.68APRR137 pKa = 11.84LIQARR142 pKa = 11.84HH143 pKa = 4.15MTHH146 pKa = 6.35NIEE149 pKa = 3.97VGRR152 pKa = 11.84YY153 pKa = 7.78LKK155 pKa = 10.4PLEE158 pKa = 4.17IAVKK162 pKa = 10.41SKK164 pKa = 10.64FINFTKK170 pKa = 10.64GNYY173 pKa = 9.63DD174 pKa = 3.18SVAARR179 pKa = 11.84IEE181 pKa = 3.99KK182 pKa = 10.03LASKK186 pKa = 9.99FKK188 pKa = 10.85YY189 pKa = 8.81FTEE192 pKa = 4.35SDD194 pKa = 3.45HH195 pKa = 5.68VTFDD199 pKa = 3.04AHH201 pKa = 5.82ITVEE205 pKa = 4.21HH206 pKa = 6.72LKK208 pKa = 8.33MTHH211 pKa = 6.38KK212 pKa = 10.68FYY214 pKa = 10.85LACFNHH220 pKa = 6.91DD221 pKa = 3.3RR222 pKa = 11.84TLQKK226 pKa = 10.68YY227 pKa = 9.31LKK229 pKa = 8.96KK230 pKa = 9.36TINNRR235 pKa = 11.84IRR237 pKa = 11.84TRR239 pKa = 11.84DD240 pKa = 3.36GTKK243 pKa = 7.21WTVRR247 pKa = 11.84GTRR250 pKa = 11.84MSGDD254 pKa = 3.13VDD256 pKa = 3.61TSFGNSLINYY266 pKa = 7.82AIIMQVLEE274 pKa = 3.82NLQLKK279 pKa = 10.19GDD281 pKa = 4.67AIVNGDD287 pKa = 3.79DD288 pKa = 5.22SIIFTNQKK296 pKa = 9.67VDD298 pKa = 3.25NKK300 pKa = 9.89LAKK303 pKa = 10.27IMFDD307 pKa = 3.19IYY309 pKa = 11.14NQEE312 pKa = 4.1TEE314 pKa = 4.12IKK316 pKa = 9.42EE317 pKa = 4.33SEE319 pKa = 4.12NSIHH323 pKa = 6.86RR324 pKa = 11.84VEE326 pKa = 4.88FCRR329 pKa = 11.84TRR331 pKa = 11.84LVYY334 pKa = 9.55PASGKK339 pKa = 7.71PTMMMDD345 pKa = 4.2PKK347 pKa = 10.22RR348 pKa = 11.84LNSIYY353 pKa = 10.92GMTYY357 pKa = 10.61KK358 pKa = 10.88LIPPEE363 pKa = 4.28EE364 pKa = 4.06YY365 pKa = 10.78ANYY368 pKa = 10.26LINIKK373 pKa = 9.93KK374 pKa = 10.53ANEE377 pKa = 4.42SINKK381 pKa = 5.87NTPIGHH387 pKa = 6.05YY388 pKa = 8.15WHH390 pKa = 6.94TDD392 pKa = 3.05EE393 pKa = 5.52EE394 pKa = 4.86VKK396 pKa = 10.38HH397 pKa = 6.68DD398 pKa = 3.53KK399 pKa = 11.05HH400 pKa = 8.0IDD402 pKa = 3.45YY403 pKa = 10.66SLKK406 pKa = 10.47RR407 pKa = 11.84VITRR411 pKa = 11.84EE412 pKa = 4.19SVNQTSTTEE421 pKa = 3.46ITITMFIAYY430 pKa = 7.67PNLLEE435 pKa = 5.12DD436 pKa = 4.18LNKK439 pKa = 8.16TTEE442 pKa = 4.56LIKK445 pKa = 10.91RR446 pKa = 11.84NLPRR450 pKa = 11.84KK451 pKa = 9.66NSPSWFIDD459 pKa = 3.1HH460 pKa = 7.56DD461 pKa = 4.27NKK463 pKa = 10.74KK464 pKa = 9.95LHH466 pKa = 5.87QLNDD470 pKa = 3.26
Molecular weight: 55.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
709 |
239 |
470 |
354.5 |
41.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.501 ± 1.066 | 0.987 ± 0.36 |
5.642 ± 0.113 | 5.783 ± 0.039 |
3.526 ± 0.126 | 2.962 ± 0.202 |
3.385 ± 0.898 | 9.873 ± 0.528 |
8.886 ± 1.15 | 6.629 ± 0.693 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.68 ± 0.309 | 7.757 ± 0.101 |
3.385 ± 0.679 | 3.949 ± 0.123 |
6.065 ± 0.111 | 5.078 ± 0.249 |
8.039 ± 1.051 | 3.808 ± 0.681 |
1.269 ± 0.432 | 4.795 ± 0.98 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
