
Agromyces atrinae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agromyces
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
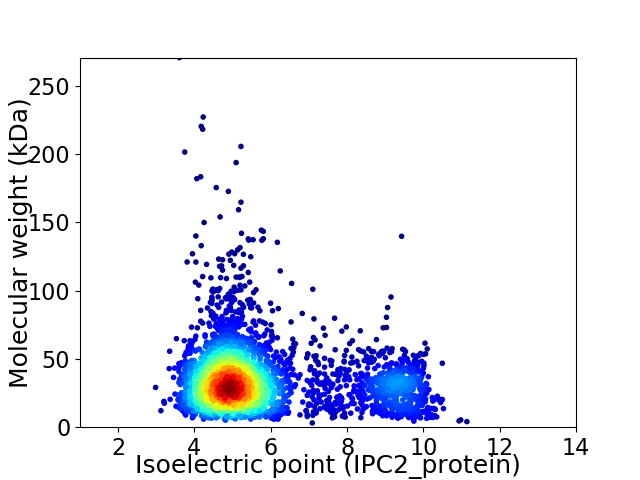
Virtual 2D-PAGE plot for 3277 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q2M775|A0A4Q2M775_9MICO DNA-(apurinic or apyrimidinic site) lyase OS=Agromyces atrinae OX=592376 GN=ESP50_13255 PE=3 SV=1
MM1 pKa = 7.18PQFRR5 pKa = 11.84NHH7 pKa = 5.26MRR9 pKa = 11.84SAGVVTAIAVAGMTLAGCASSSEE32 pKa = 4.24STEE35 pKa = 3.84AGGEE39 pKa = 4.17KK40 pKa = 8.25ITYY43 pKa = 7.77WLWDD47 pKa = 3.71TNQQPAYY54 pKa = 6.96TQCATAFTEE63 pKa = 4.06KK64 pKa = 10.14TGIDD68 pKa = 3.6VTIEE72 pKa = 3.72QYY74 pKa = 10.81GWADD78 pKa = 3.15YY79 pKa = 9.36WQGLTTGFAAGTAPDD94 pKa = 3.68VFADD98 pKa = 3.48HH99 pKa = 7.13LSYY102 pKa = 11.07YY103 pKa = 9.22PQFVSQGQLLDD114 pKa = 3.21ISDD117 pKa = 5.27RR118 pKa = 11.84IDD120 pKa = 3.58ADD122 pKa = 4.6GIDD125 pKa = 3.57LSIYY129 pKa = 10.29QDD131 pKa = 3.43GLADD135 pKa = 3.8LWVDD139 pKa = 3.72QEE141 pKa = 4.16GGRR144 pKa = 11.84YY145 pKa = 9.09GLPKK149 pKa = 10.9DD150 pKa = 3.91FDD152 pKa = 4.36TVALFYY158 pKa = 11.12NEE160 pKa = 4.12TMVTDD165 pKa = 4.22AGIDD169 pKa = 3.49PAALSEE175 pKa = 4.31AAWNPTDD182 pKa = 4.47GGTFEE187 pKa = 4.17QAVAALTVDD196 pKa = 3.63EE197 pKa = 4.56NGVRR201 pKa = 11.84GTEE204 pKa = 4.16AGFDD208 pKa = 3.72KK209 pKa = 11.43NNVAVYY215 pKa = 10.39GLALSGNGLNASGQQTWAPYY235 pKa = 11.08ALGNDD240 pKa = 3.62WYY242 pKa = 11.09FGDD245 pKa = 3.77TTPWTTEE252 pKa = 3.67WNFDD256 pKa = 3.48APEE259 pKa = 4.16FTDD262 pKa = 3.19TLAWYY267 pKa = 10.07KK268 pKa = 10.69SLQDD272 pKa = 3.43KK273 pKa = 11.09GYY275 pKa = 9.42MPTIDD280 pKa = 3.69IALSEE285 pKa = 4.04QDD287 pKa = 3.98PLNGYY292 pKa = 9.07LAGRR296 pKa = 11.84YY297 pKa = 9.5ALVTDD302 pKa = 4.86GSWMNSSYY310 pKa = 10.91LGQSDD315 pKa = 4.56VPTKK319 pKa = 10.49VAPTPVGPSGEE330 pKa = 4.09RR331 pKa = 11.84ASVFNGLSDD340 pKa = 5.16GIWAGTKK347 pKa = 10.53NPDD350 pKa = 4.32AAWEE354 pKa = 4.26WVSYY358 pKa = 10.12LASTDD363 pKa = 3.82CQDD366 pKa = 3.53VVADD370 pKa = 3.86AAVVFPAIKK379 pKa = 9.83TSTEE383 pKa = 3.65KK384 pKa = 10.31ATEE387 pKa = 3.81AFAAKK392 pKa = 9.72GWDD395 pKa = 3.29VSGFAVQVDD404 pKa = 4.08EE405 pKa = 4.72GTTALLPIADD415 pKa = 4.15HH416 pKa = 6.21WSEE419 pKa = 4.2INDD422 pKa = 3.54LVTANSQAFLKK433 pKa = 10.85GSVDD437 pKa = 3.4AEE439 pKa = 4.45SFTGVNDD446 pKa = 3.39QVNALFQQ453 pKa = 4.05
MM1 pKa = 7.18PQFRR5 pKa = 11.84NHH7 pKa = 5.26MRR9 pKa = 11.84SAGVVTAIAVAGMTLAGCASSSEE32 pKa = 4.24STEE35 pKa = 3.84AGGEE39 pKa = 4.17KK40 pKa = 8.25ITYY43 pKa = 7.77WLWDD47 pKa = 3.71TNQQPAYY54 pKa = 6.96TQCATAFTEE63 pKa = 4.06KK64 pKa = 10.14TGIDD68 pKa = 3.6VTIEE72 pKa = 3.72QYY74 pKa = 10.81GWADD78 pKa = 3.15YY79 pKa = 9.36WQGLTTGFAAGTAPDD94 pKa = 3.68VFADD98 pKa = 3.48HH99 pKa = 7.13LSYY102 pKa = 11.07YY103 pKa = 9.22PQFVSQGQLLDD114 pKa = 3.21ISDD117 pKa = 5.27RR118 pKa = 11.84IDD120 pKa = 3.58ADD122 pKa = 4.6GIDD125 pKa = 3.57LSIYY129 pKa = 10.29QDD131 pKa = 3.43GLADD135 pKa = 3.8LWVDD139 pKa = 3.72QEE141 pKa = 4.16GGRR144 pKa = 11.84YY145 pKa = 9.09GLPKK149 pKa = 10.9DD150 pKa = 3.91FDD152 pKa = 4.36TVALFYY158 pKa = 11.12NEE160 pKa = 4.12TMVTDD165 pKa = 4.22AGIDD169 pKa = 3.49PAALSEE175 pKa = 4.31AAWNPTDD182 pKa = 4.47GGTFEE187 pKa = 4.17QAVAALTVDD196 pKa = 3.63EE197 pKa = 4.56NGVRR201 pKa = 11.84GTEE204 pKa = 4.16AGFDD208 pKa = 3.72KK209 pKa = 11.43NNVAVYY215 pKa = 10.39GLALSGNGLNASGQQTWAPYY235 pKa = 11.08ALGNDD240 pKa = 3.62WYY242 pKa = 11.09FGDD245 pKa = 3.77TTPWTTEE252 pKa = 3.67WNFDD256 pKa = 3.48APEE259 pKa = 4.16FTDD262 pKa = 3.19TLAWYY267 pKa = 10.07KK268 pKa = 10.69SLQDD272 pKa = 3.43KK273 pKa = 11.09GYY275 pKa = 9.42MPTIDD280 pKa = 3.69IALSEE285 pKa = 4.04QDD287 pKa = 3.98PLNGYY292 pKa = 9.07LAGRR296 pKa = 11.84YY297 pKa = 9.5ALVTDD302 pKa = 4.86GSWMNSSYY310 pKa = 10.91LGQSDD315 pKa = 4.56VPTKK319 pKa = 10.49VAPTPVGPSGEE330 pKa = 4.09RR331 pKa = 11.84ASVFNGLSDD340 pKa = 5.16GIWAGTKK347 pKa = 10.53NPDD350 pKa = 4.32AAWEE354 pKa = 4.26WVSYY358 pKa = 10.12LASTDD363 pKa = 3.82CQDD366 pKa = 3.53VVADD370 pKa = 3.86AAVVFPAIKK379 pKa = 9.83TSTEE383 pKa = 3.65KK384 pKa = 10.31ATEE387 pKa = 3.81AFAAKK392 pKa = 9.72GWDD395 pKa = 3.29VSGFAVQVDD404 pKa = 4.08EE405 pKa = 4.72GTTALLPIADD415 pKa = 4.15HH416 pKa = 6.21WSEE419 pKa = 4.2INDD422 pKa = 3.54LVTANSQAFLKK433 pKa = 10.85GSVDD437 pKa = 3.4AEE439 pKa = 4.45SFTGVNDD446 pKa = 3.39QVNALFQQ453 pKa = 4.05
Molecular weight: 48.63 kDa
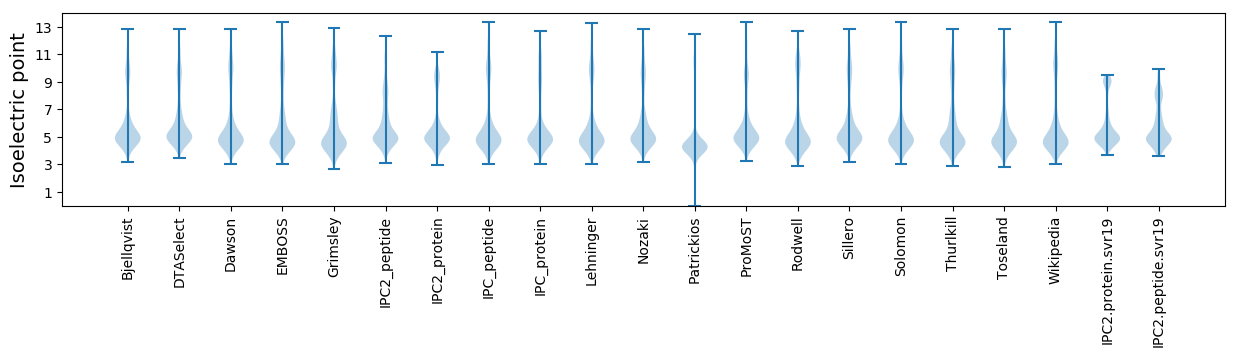
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q2M4L5|A0A4Q2M4L5_9MICO Pyruvate dehydrogenase (Acetyl-transferring) E1 component subunit alpha OS=Agromyces atrinae OX=592376 GN=pdhA PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
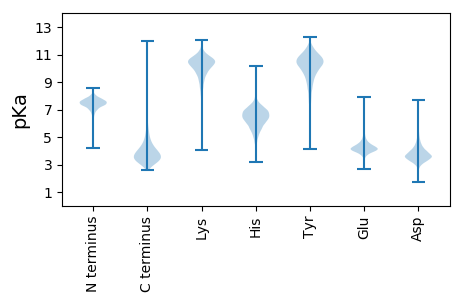
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1105242 |
29 |
2641 |
337.3 |
36.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.423 ± 0.061 | 0.451 ± 0.008 |
6.423 ± 0.038 | 5.722 ± 0.039 |
3.293 ± 0.027 | 8.879 ± 0.036 |
1.874 ± 0.02 | 4.874 ± 0.037 |
1.771 ± 0.028 | 10.102 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.558 ± 0.015 | 1.996 ± 0.024 |
5.27 ± 0.029 | 2.376 ± 0.02 |
7.189 ± 0.048 | 6.06 ± 0.032 |
6.16 ± 0.039 | 9.104 ± 0.04 |
1.486 ± 0.02 | 1.988 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
