
Porcine reproductive and respiratory syndrome virus (strain Lelystad) (PRRSV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Arnidovirineae; Arteriviridae; Variarterivirinae; Betaarterivirus; Eurpobartevirus; Betaarterivirus suid 1; Porcine reproductive and respiratory syndrome virus 1
Average proteome isoelectric point is 7.66
Get precalculated fractions of proteins
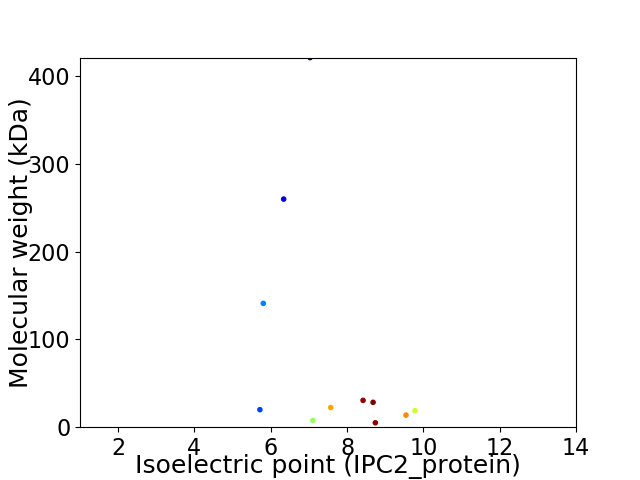
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
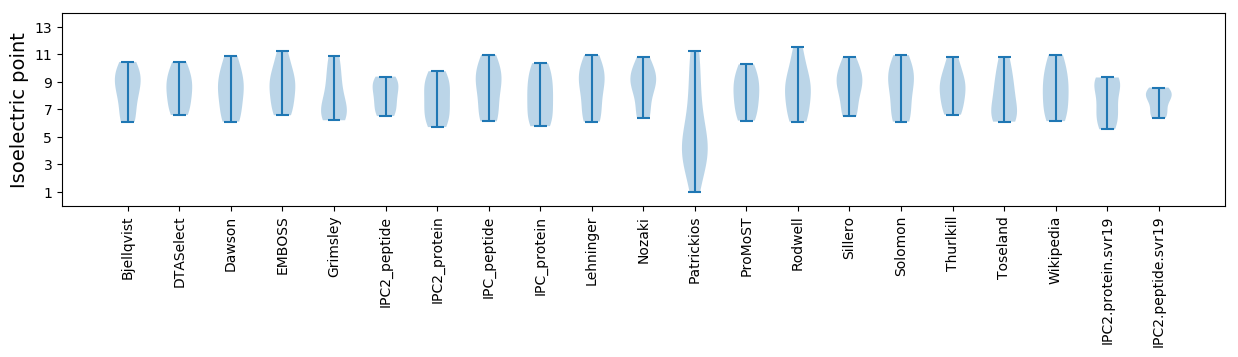
* You can choose from 21 different methods for calculating isoelectric point
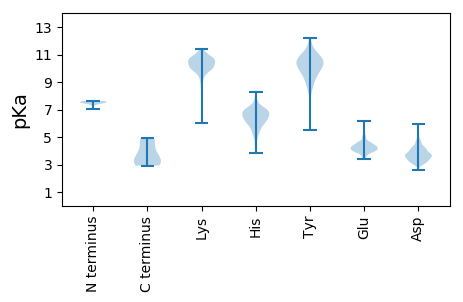
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q04569|GP5_PRRSL Glycoprotein 5 OS=Porcine reproductive and respiratory syndrome virus (strain Lelystad) OX=11049 GN=GP5 PE=1 SV=1
MM1 pKa = 7.52AAATLFFLAGAQHH14 pKa = 6.09IMVSEE19 pKa = 4.3AFACKK24 pKa = 9.81PCFSTHH30 pKa = 6.63LSDD33 pKa = 5.1IEE35 pKa = 4.57TNTTAAAGFMVLQDD49 pKa = 3.42INCFRR54 pKa = 11.84PHH56 pKa = 6.1GVSAAQEE63 pKa = 4.18KK64 pKa = 10.51ISFGKK69 pKa = 10.43SSQCRR74 pKa = 11.84EE75 pKa = 4.07AVGTPQYY82 pKa = 10.07ITITANVTDD91 pKa = 4.07EE92 pKa = 4.52SYY94 pKa = 11.16LYY96 pKa = 10.94NADD99 pKa = 4.24LLMLSACLFYY109 pKa = 10.88ASEE112 pKa = 4.12MSEE115 pKa = 4.0KK116 pKa = 10.14GFKK119 pKa = 10.72VIFGNVSGVVSACVNFTDD137 pKa = 4.45YY138 pKa = 10.78VAHH141 pKa = 5.73VTQHH145 pKa = 5.01TQQHH149 pKa = 6.36HH150 pKa = 5.87LVIDD154 pKa = 4.64HH155 pKa = 6.7IRR157 pKa = 11.84LLHH160 pKa = 6.26FLTPSAMRR168 pKa = 11.84WATTIACLFAILLAII183 pKa = 4.72
MM1 pKa = 7.52AAATLFFLAGAQHH14 pKa = 6.09IMVSEE19 pKa = 4.3AFACKK24 pKa = 9.81PCFSTHH30 pKa = 6.63LSDD33 pKa = 5.1IEE35 pKa = 4.57TNTTAAAGFMVLQDD49 pKa = 3.42INCFRR54 pKa = 11.84PHH56 pKa = 6.1GVSAAQEE63 pKa = 4.18KK64 pKa = 10.51ISFGKK69 pKa = 10.43SSQCRR74 pKa = 11.84EE75 pKa = 4.07AVGTPQYY82 pKa = 10.07ITITANVTDD91 pKa = 4.07EE92 pKa = 4.52SYY94 pKa = 11.16LYY96 pKa = 10.94NADD99 pKa = 4.24LLMLSACLFYY109 pKa = 10.88ASEE112 pKa = 4.12MSEE115 pKa = 4.0KK116 pKa = 10.14GFKK119 pKa = 10.72VIFGNVSGVVSACVNFTDD137 pKa = 4.45YY138 pKa = 10.78VAHH141 pKa = 5.73VTQHH145 pKa = 5.01TQQHH149 pKa = 6.36HH150 pKa = 5.87LVIDD154 pKa = 4.64HH155 pKa = 6.7IRR157 pKa = 11.84LLHH160 pKa = 6.26FLTPSAMRR168 pKa = 11.84WATTIACLFAILLAII183 pKa = 4.72
Molecular weight: 19.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q04566|GP2A_PRRSL Glycoprotein 2a OS=Porcine reproductive and respiratory syndrome virus (strain Lelystad) OX=11049 GN=GP2a PE=1 SV=1
MM1 pKa = 7.6GGLDD5 pKa = 4.73DD6 pKa = 5.03FCNDD10 pKa = 4.64PIAAQKK16 pKa = 10.6LVLAFSITYY25 pKa = 7.79TPIMIYY31 pKa = 10.21ALKK34 pKa = 10.28VSRR37 pKa = 11.84GRR39 pKa = 11.84LLGLLHH45 pKa = 7.33ILIFLNCSFTFGYY58 pKa = 6.87MTYY61 pKa = 10.53VHH63 pKa = 6.61FQSTNRR69 pKa = 11.84VALTLGAVVALLWGVYY85 pKa = 10.17SFTEE89 pKa = 3.64SWKK92 pKa = 10.78FITSRR97 pKa = 11.84CRR99 pKa = 11.84LCCLGRR105 pKa = 11.84RR106 pKa = 11.84YY107 pKa = 9.92ILAPAHH113 pKa = 6.09HH114 pKa = 6.83VEE116 pKa = 4.56SAAGLHH122 pKa = 6.44SISASGNRR130 pKa = 11.84AYY132 pKa = 10.44AVRR135 pKa = 11.84KK136 pKa = 9.38PGLTSVNGTLVPGLRR151 pKa = 11.84SLVLGGKK158 pKa = 9.24RR159 pKa = 11.84AVKK162 pKa = 10.05RR163 pKa = 11.84GVVNLVKK170 pKa = 10.72YY171 pKa = 10.64GRR173 pKa = 3.87
MM1 pKa = 7.6GGLDD5 pKa = 4.73DD6 pKa = 5.03FCNDD10 pKa = 4.64PIAAQKK16 pKa = 10.6LVLAFSITYY25 pKa = 7.79TPIMIYY31 pKa = 10.21ALKK34 pKa = 10.28VSRR37 pKa = 11.84GRR39 pKa = 11.84LLGLLHH45 pKa = 7.33ILIFLNCSFTFGYY58 pKa = 6.87MTYY61 pKa = 10.53VHH63 pKa = 6.61FQSTNRR69 pKa = 11.84VALTLGAVVALLWGVYY85 pKa = 10.17SFTEE89 pKa = 3.64SWKK92 pKa = 10.78FITSRR97 pKa = 11.84CRR99 pKa = 11.84LCCLGRR105 pKa = 11.84RR106 pKa = 11.84YY107 pKa = 9.92ILAPAHH113 pKa = 6.09HH114 pKa = 6.83VEE116 pKa = 4.56SAAGLHH122 pKa = 6.44SISASGNRR130 pKa = 11.84AYY132 pKa = 10.44AVRR135 pKa = 11.84KK136 pKa = 9.38PGLTSVNGTLVPGLRR151 pKa = 11.84SLVLGGKK158 pKa = 9.24RR159 pKa = 11.84AVKK162 pKa = 10.05RR163 pKa = 11.84GVVNLVKK170 pKa = 10.72YY171 pKa = 10.64GRR173 pKa = 3.87
Molecular weight: 18.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8848 |
43 |
3855 |
804.4 |
88.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.544 ± 0.27 | 3.052 ± 0.083 |
4.713 ± 0.423 | 4.204 ± 0.31 |
4.509 ± 0.512 | 7.177 ± 0.333 |
2.633 ± 0.278 | 3.617 ± 0.347 |
3.922 ± 0.309 | 10.533 ± 0.276 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.763 ± 0.102 | 2.735 ± 0.085 |
6.939 ± 0.78 | 3.639 ± 0.271 |
5.448 ± 0.217 | 8.262 ± 0.493 |
5.741 ± 0.29 | 8.16 ± 0.195 |
1.989 ± 0.246 | 2.419 ± 0.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
