
Fonticula alba (Slime mold)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Rotosphaerida; Fonticulaceae; Fonticula
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
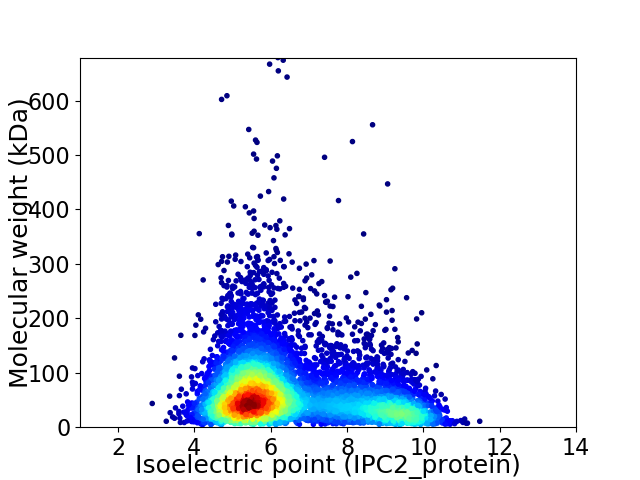
Virtual 2D-PAGE plot for 6244 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A058ZDV0|A0A058ZDV0_FONAL VPS11_C domain-containing protein OS=Fonticula alba OX=691883 GN=H696_01059 PE=3 SV=1
MM1 pKa = 7.56ASSLFRR7 pKa = 11.84AAPLMADD14 pKa = 4.45TFTQAQPPVAPGTEE28 pKa = 4.19QSAMPAQSAPPAHH41 pKa = 6.7PTSMPQTPGTPPPALVDD58 pKa = 3.55PFFSGRR64 pKa = 11.84GPTGQEE70 pKa = 3.14ADD72 pKa = 3.83LLGEE76 pKa = 4.02GLVTHH81 pKa = 6.59EE82 pKa = 4.58AHH84 pKa = 7.65DD85 pKa = 4.8DD86 pKa = 3.65GAGAFDD92 pKa = 5.63DD93 pKa = 4.3GRR95 pKa = 11.84LDD97 pKa = 4.76ARR99 pKa = 11.84PEE101 pKa = 3.9DD102 pKa = 4.2LGADD106 pKa = 4.02GDD108 pKa = 5.55DD109 pKa = 6.03DD110 pKa = 7.02DD111 pKa = 7.76DD112 pKa = 7.72DD113 pKa = 7.61DD114 pKa = 7.68DD115 pKa = 7.64DD116 pKa = 7.68DD117 pKa = 7.61DD118 pKa = 7.65DD119 pKa = 7.0DD120 pKa = 5.71DD121 pKa = 5.74LAMTPRR127 pKa = 11.84VAAQFYY133 pKa = 10.12LPQIYY138 pKa = 10.47GLFASLFATFFLVIWWAPTIFLSVPFNYY166 pKa = 9.9TGVVLSTGTNTLSTGSQLLVAVASAAILLVIILVNTVILLALFYY210 pKa = 10.35YY211 pKa = 10.49GYY213 pKa = 10.51IRR215 pKa = 11.84VIYY218 pKa = 9.46GWLIVATGLILSFIGGIVFYY238 pKa = 10.8RR239 pKa = 11.84LLEE242 pKa = 4.37TYY244 pKa = 9.9NLPVDD249 pKa = 3.47WVTFCFLLFNFSVVGLLAVFGKK271 pKa = 8.77LTLRR275 pKa = 11.84MQQVYY280 pKa = 9.79LVIVSALVATNLSGLPSWTAWAVLIVVSIYY310 pKa = 11.18DD311 pKa = 4.26LFAVLCPQGPLNLLINLSQTRR332 pKa = 11.84QQSIPALLYY341 pKa = 8.77SAGMARR347 pKa = 11.84LASSNDD353 pKa = 3.1DD354 pKa = 3.66SVPILAFTAGTSDD367 pKa = 4.67DD368 pKa = 4.7SGDD371 pKa = 3.99EE372 pKa = 4.05QPEE375 pKa = 4.13DD376 pKa = 4.07ADD378 pKa = 4.14TQVGQALLSSGHH390 pKa = 6.62LEE392 pKa = 4.05SVLLGEE398 pKa = 5.12AMPSAASDD406 pKa = 3.84LDD408 pKa = 4.05SAAVPAIDD416 pKa = 3.9STTGNPILDD425 pKa = 4.26AEE427 pKa = 4.58TGPKK431 pKa = 9.96DD432 pKa = 4.17VSDD435 pKa = 4.06EE436 pKa = 4.1QDD438 pKa = 4.88DD439 pKa = 4.45GDD441 pKa = 5.8DD442 pKa = 5.33DD443 pKa = 5.35DD444 pKa = 7.07GEE446 pKa = 6.33DD447 pKa = 3.78GDD449 pKa = 4.54EE450 pKa = 4.35PPGRR454 pKa = 11.84GPVQLGLGDD463 pKa = 4.11YY464 pKa = 10.34IFYY467 pKa = 10.74SLLVARR473 pKa = 11.84GALTSDD479 pKa = 3.65GSDD482 pKa = 3.08GGASAAIAVISCVLSTLVGLGLTLSLLALFRR513 pKa = 11.84KK514 pKa = 8.81PLPALPISIGLGILAYY530 pKa = 10.38VFSAFLSTPMVVEE543 pKa = 4.12MTRR546 pKa = 11.84QVVFVV551 pKa = 3.87
MM1 pKa = 7.56ASSLFRR7 pKa = 11.84AAPLMADD14 pKa = 4.45TFTQAQPPVAPGTEE28 pKa = 4.19QSAMPAQSAPPAHH41 pKa = 6.7PTSMPQTPGTPPPALVDD58 pKa = 3.55PFFSGRR64 pKa = 11.84GPTGQEE70 pKa = 3.14ADD72 pKa = 3.83LLGEE76 pKa = 4.02GLVTHH81 pKa = 6.59EE82 pKa = 4.58AHH84 pKa = 7.65DD85 pKa = 4.8DD86 pKa = 3.65GAGAFDD92 pKa = 5.63DD93 pKa = 4.3GRR95 pKa = 11.84LDD97 pKa = 4.76ARR99 pKa = 11.84PEE101 pKa = 3.9DD102 pKa = 4.2LGADD106 pKa = 4.02GDD108 pKa = 5.55DD109 pKa = 6.03DD110 pKa = 7.02DD111 pKa = 7.76DD112 pKa = 7.72DD113 pKa = 7.61DD114 pKa = 7.68DD115 pKa = 7.64DD116 pKa = 7.68DD117 pKa = 7.61DD118 pKa = 7.65DD119 pKa = 7.0DD120 pKa = 5.71DD121 pKa = 5.74LAMTPRR127 pKa = 11.84VAAQFYY133 pKa = 10.12LPQIYY138 pKa = 10.47GLFASLFATFFLVIWWAPTIFLSVPFNYY166 pKa = 9.9TGVVLSTGTNTLSTGSQLLVAVASAAILLVIILVNTVILLALFYY210 pKa = 10.35YY211 pKa = 10.49GYY213 pKa = 10.51IRR215 pKa = 11.84VIYY218 pKa = 9.46GWLIVATGLILSFIGGIVFYY238 pKa = 10.8RR239 pKa = 11.84LLEE242 pKa = 4.37TYY244 pKa = 9.9NLPVDD249 pKa = 3.47WVTFCFLLFNFSVVGLLAVFGKK271 pKa = 8.77LTLRR275 pKa = 11.84MQQVYY280 pKa = 9.79LVIVSALVATNLSGLPSWTAWAVLIVVSIYY310 pKa = 11.18DD311 pKa = 4.26LFAVLCPQGPLNLLINLSQTRR332 pKa = 11.84QQSIPALLYY341 pKa = 8.77SAGMARR347 pKa = 11.84LASSNDD353 pKa = 3.1DD354 pKa = 3.66SVPILAFTAGTSDD367 pKa = 4.67DD368 pKa = 4.7SGDD371 pKa = 3.99EE372 pKa = 4.05QPEE375 pKa = 4.13DD376 pKa = 4.07ADD378 pKa = 4.14TQVGQALLSSGHH390 pKa = 6.62LEE392 pKa = 4.05SVLLGEE398 pKa = 5.12AMPSAASDD406 pKa = 3.84LDD408 pKa = 4.05SAAVPAIDD416 pKa = 3.9STTGNPILDD425 pKa = 4.26AEE427 pKa = 4.58TGPKK431 pKa = 9.96DD432 pKa = 4.17VSDD435 pKa = 4.06EE436 pKa = 4.1QDD438 pKa = 4.88DD439 pKa = 4.45GDD441 pKa = 5.8DD442 pKa = 5.33DD443 pKa = 5.35DD444 pKa = 7.07GEE446 pKa = 6.33DD447 pKa = 3.78GDD449 pKa = 4.54EE450 pKa = 4.35PPGRR454 pKa = 11.84GPVQLGLGDD463 pKa = 4.11YY464 pKa = 10.34IFYY467 pKa = 10.74SLLVARR473 pKa = 11.84GALTSDD479 pKa = 3.65GSDD482 pKa = 3.08GGASAAIAVISCVLSTLVGLGLTLSLLALFRR513 pKa = 11.84KK514 pKa = 8.81PLPALPISIGLGILAYY530 pKa = 10.38VFSAFLSTPMVVEE543 pKa = 4.12MTRR546 pKa = 11.84QVVFVV551 pKa = 3.87
Molecular weight: 58.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A058Z4E6|A0A058Z4E6_FONAL Uncharacterized protein OS=Fonticula alba OX=691883 GN=H696_04391 PE=4 SV=1
MM1 pKa = 7.53RR2 pKa = 11.84SAWSSRR8 pKa = 11.84RR9 pKa = 11.84SASRR13 pKa = 11.84EE14 pKa = 3.57GSFIAPAAGSAPAGPPPMRR33 pKa = 11.84LRR35 pKa = 11.84RR36 pKa = 11.84IRR38 pKa = 11.84PPRR41 pKa = 11.84PRR43 pKa = 11.84PRR45 pKa = 11.84LLRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84PLAPAIPLGPSRR66 pKa = 4.73
MM1 pKa = 7.53RR2 pKa = 11.84SAWSSRR8 pKa = 11.84RR9 pKa = 11.84SASRR13 pKa = 11.84EE14 pKa = 3.57GSFIAPAAGSAPAGPPPMRR33 pKa = 11.84LRR35 pKa = 11.84RR36 pKa = 11.84IRR38 pKa = 11.84PPRR41 pKa = 11.84PRR43 pKa = 11.84PRR45 pKa = 11.84LLRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84PLAPAIPLGPSRR66 pKa = 4.73
Molecular weight: 7.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4258807 |
42 |
6752 |
682.1 |
71.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.272 ± 0.054 | 1.723 ± 0.036 |
5.239 ± 0.024 | 4.49 ± 0.027 |
3.144 ± 0.018 | 8.784 ± 0.049 |
2.683 ± 0.019 | 3.126 ± 0.024 |
2.001 ± 0.021 | 9.905 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.905 ± 0.011 | 2.068 ± 0.017 |
8.675 ± 0.051 | 3.668 ± 0.027 |
6.665 ± 0.026 | 9.101 ± 0.048 |
5.177 ± 0.017 | 5.723 ± 0.022 |
0.937 ± 0.008 | 1.712 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
