
Thrips-associated genomovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; unclassified Genomoviridae
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
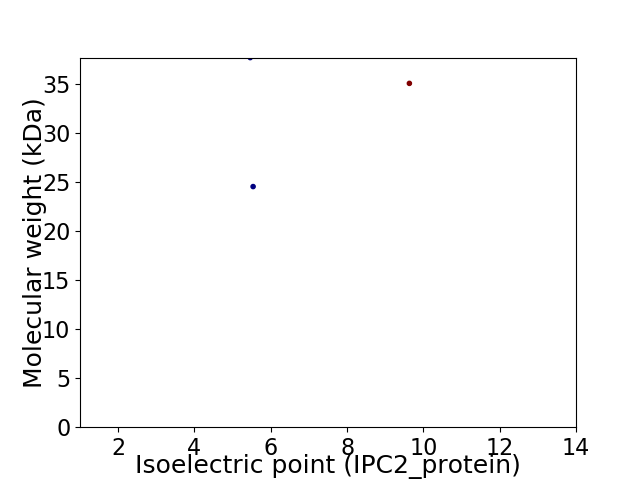
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8YT76|A0A1P8YT76_9VIRU RepA OS=Thrips-associated genomovirus 2 OX=1941234 PE=3 SV=1
MM1 pKa = 7.47TFLFSARR8 pKa = 11.84YY9 pKa = 9.42VLLTYY14 pKa = 9.1AQCGTLDD21 pKa = 3.17GWAVSDD27 pKa = 4.02HH28 pKa = 6.52LSALGAEE35 pKa = 4.64CIVGRR40 pKa = 11.84EE41 pKa = 4.0NHH43 pKa = 6.3SDD45 pKa = 3.41GGTHH49 pKa = 6.32LHH51 pKa = 6.56AFVDD55 pKa = 4.89FGRR58 pKa = 11.84KK59 pKa = 8.11KK60 pKa = 9.9QSRR63 pKa = 11.84RR64 pKa = 11.84PDD66 pKa = 3.25FFDD69 pKa = 4.06VGGHH73 pKa = 6.35HH74 pKa = 7.14PNIAPSRR81 pKa = 11.84GRR83 pKa = 11.84PEE85 pKa = 3.88RR86 pKa = 11.84GYY88 pKa = 11.11DD89 pKa = 3.48YY90 pKa = 10.71AIKK93 pKa = 10.87DD94 pKa = 3.59GDD96 pKa = 4.02VVAGGLARR104 pKa = 11.84PGGGGLPEE112 pKa = 4.34VANKK116 pKa = 9.25WSEE119 pKa = 3.81IVAAEE124 pKa = 4.05SRR126 pKa = 11.84DD127 pKa = 3.63EE128 pKa = 4.87FFDD131 pKa = 5.05LLRR134 pKa = 11.84QLDD137 pKa = 4.12PKK139 pKa = 10.46TLVTRR144 pKa = 11.84WTEE147 pKa = 3.7LNKK150 pKa = 10.8YY151 pKa = 10.14ADD153 pKa = 3.7AAYY156 pKa = 10.25APTPEE161 pKa = 5.03PYY163 pKa = 10.25VGSDD167 pKa = 2.91GKK169 pKa = 10.54QFEE172 pKa = 4.79LGMVPEE178 pKa = 4.41LARR181 pKa = 11.84WGEE184 pKa = 3.83QLVANDD190 pKa = 4.69PIEE193 pKa = 4.03GRR195 pKa = 11.84MRR197 pKa = 11.84SLVLYY202 pKa = 10.3GPSRR206 pKa = 11.84LGKK209 pKa = 8.13TIWARR214 pKa = 11.84SLGPHH219 pKa = 6.3VYY221 pKa = 11.09SMGIVSGKK229 pKa = 10.23LLLRR233 pKa = 11.84DD234 pKa = 3.77APGAAYY240 pKa = 9.98AVFDD244 pKa = 4.61DD245 pKa = 3.64MRR247 pKa = 11.84GGIGYY252 pKa = 7.9FHH254 pKa = 7.09SWKK257 pKa = 9.79EE258 pKa = 3.68WLGAQSVVTVKK269 pKa = 10.33EE270 pKa = 4.2LYY272 pKa = 9.8RR273 pKa = 11.84DD274 pKa = 3.61PVQLVWGRR282 pKa = 11.84PCIWLANRR290 pKa = 11.84DD291 pKa = 3.63PRR293 pKa = 11.84LEE295 pKa = 3.98LWADD299 pKa = 3.46LTDD302 pKa = 4.15RR303 pKa = 11.84SAASKK308 pKa = 10.82RR309 pKa = 11.84DD310 pKa = 3.8MIQSDD315 pKa = 4.44VDD317 pKa = 3.37WLEE320 pKa = 4.03ANCIFVEE327 pKa = 4.16LQEE330 pKa = 4.74PIFRR334 pKa = 11.84ANTEE338 pKa = 3.84
MM1 pKa = 7.47TFLFSARR8 pKa = 11.84YY9 pKa = 9.42VLLTYY14 pKa = 9.1AQCGTLDD21 pKa = 3.17GWAVSDD27 pKa = 4.02HH28 pKa = 6.52LSALGAEE35 pKa = 4.64CIVGRR40 pKa = 11.84EE41 pKa = 4.0NHH43 pKa = 6.3SDD45 pKa = 3.41GGTHH49 pKa = 6.32LHH51 pKa = 6.56AFVDD55 pKa = 4.89FGRR58 pKa = 11.84KK59 pKa = 8.11KK60 pKa = 9.9QSRR63 pKa = 11.84RR64 pKa = 11.84PDD66 pKa = 3.25FFDD69 pKa = 4.06VGGHH73 pKa = 6.35HH74 pKa = 7.14PNIAPSRR81 pKa = 11.84GRR83 pKa = 11.84PEE85 pKa = 3.88RR86 pKa = 11.84GYY88 pKa = 11.11DD89 pKa = 3.48YY90 pKa = 10.71AIKK93 pKa = 10.87DD94 pKa = 3.59GDD96 pKa = 4.02VVAGGLARR104 pKa = 11.84PGGGGLPEE112 pKa = 4.34VANKK116 pKa = 9.25WSEE119 pKa = 3.81IVAAEE124 pKa = 4.05SRR126 pKa = 11.84DD127 pKa = 3.63EE128 pKa = 4.87FFDD131 pKa = 5.05LLRR134 pKa = 11.84QLDD137 pKa = 4.12PKK139 pKa = 10.46TLVTRR144 pKa = 11.84WTEE147 pKa = 3.7LNKK150 pKa = 10.8YY151 pKa = 10.14ADD153 pKa = 3.7AAYY156 pKa = 10.25APTPEE161 pKa = 5.03PYY163 pKa = 10.25VGSDD167 pKa = 2.91GKK169 pKa = 10.54QFEE172 pKa = 4.79LGMVPEE178 pKa = 4.41LARR181 pKa = 11.84WGEE184 pKa = 3.83QLVANDD190 pKa = 4.69PIEE193 pKa = 4.03GRR195 pKa = 11.84MRR197 pKa = 11.84SLVLYY202 pKa = 10.3GPSRR206 pKa = 11.84LGKK209 pKa = 8.13TIWARR214 pKa = 11.84SLGPHH219 pKa = 6.3VYY221 pKa = 11.09SMGIVSGKK229 pKa = 10.23LLLRR233 pKa = 11.84DD234 pKa = 3.77APGAAYY240 pKa = 9.98AVFDD244 pKa = 4.61DD245 pKa = 3.64MRR247 pKa = 11.84GGIGYY252 pKa = 7.9FHH254 pKa = 7.09SWKK257 pKa = 9.79EE258 pKa = 3.68WLGAQSVVTVKK269 pKa = 10.33EE270 pKa = 4.2LYY272 pKa = 9.8RR273 pKa = 11.84DD274 pKa = 3.61PVQLVWGRR282 pKa = 11.84PCIWLANRR290 pKa = 11.84DD291 pKa = 3.63PRR293 pKa = 11.84LEE295 pKa = 3.98LWADD299 pKa = 3.46LTDD302 pKa = 4.15RR303 pKa = 11.84SAASKK308 pKa = 10.82RR309 pKa = 11.84DD310 pKa = 3.8MIQSDD315 pKa = 4.44VDD317 pKa = 3.37WLEE320 pKa = 4.03ANCIFVEE327 pKa = 4.16LQEE330 pKa = 4.74PIFRR334 pKa = 11.84ANTEE338 pKa = 3.84
Molecular weight: 37.69 kDa
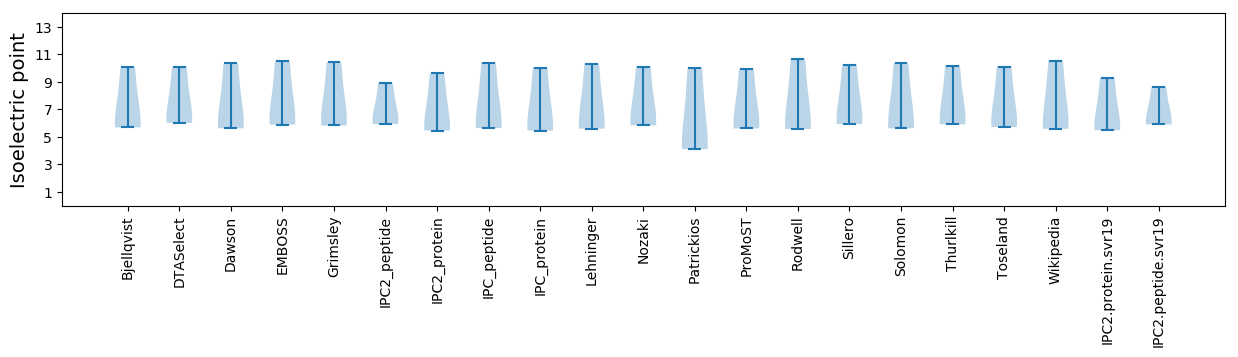
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8YT75|A0A1P8YT75_9VIRU Replication associated protein OS=Thrips-associated genomovirus 2 OX=1941234 PE=3 SV=1
MM1 pKa = 7.89PYY3 pKa = 10.18LRR5 pKa = 11.84KK6 pKa = 9.48RR7 pKa = 11.84RR8 pKa = 11.84YY9 pKa = 9.05ARR11 pKa = 11.84PRR13 pKa = 11.84YY14 pKa = 9.17SSYY17 pKa = 9.43RR18 pKa = 11.84AKK20 pKa = 10.52RR21 pKa = 11.84RR22 pKa = 11.84TPSSRR27 pKa = 11.84APYY30 pKa = 9.19RR31 pKa = 11.84YY32 pKa = 9.34SRR34 pKa = 11.84KK35 pKa = 8.56RR36 pKa = 11.84TISRR40 pKa = 11.84PRR42 pKa = 11.84YY43 pKa = 8.07AMSRR47 pKa = 11.84KK48 pKa = 9.16RR49 pKa = 11.84LLNITSRR56 pKa = 11.84KK57 pKa = 9.19KK58 pKa = 10.18QDD60 pKa = 3.04TMLVNTNTVAGTPFGGTTFSNTAALLQGGNTYY92 pKa = 10.35IFPWVATARR101 pKa = 11.84DD102 pKa = 3.94NTIGTASGNVFNSADD117 pKa = 3.76RR118 pKa = 11.84TATTCYY124 pKa = 9.66MRR126 pKa = 11.84GLKK129 pKa = 9.81EE130 pKa = 4.23RR131 pKa = 11.84IQIQTNTGMPWQWRR145 pKa = 11.84RR146 pKa = 11.84ICFTMKK152 pKa = 10.67GDD154 pKa = 6.5DD155 pKa = 3.21INAWNEE161 pKa = 3.91SAYY164 pKa = 9.95KK165 pKa = 10.41LQLEE169 pKa = 4.8TSNGWARR176 pKa = 11.84VVSNTGVASNLYY188 pKa = 9.9NNLVPLLFKK197 pKa = 10.7GAQNVDD203 pKa = 2.78WNNPFNAKK211 pKa = 8.72TDD213 pKa = 3.57NTRR216 pKa = 11.84LSIKK220 pKa = 9.94YY221 pKa = 10.31DD222 pKa = 2.75KK223 pKa = 7.68TTPIRR228 pKa = 11.84CGNSNGMMKK237 pKa = 10.28EE238 pKa = 4.27FIRR241 pKa = 11.84WHH243 pKa = 6.23PMNKK247 pKa = 9.58NLVYY251 pKa = 10.83DD252 pKa = 4.27DD253 pKa = 5.28DD254 pKa = 4.83EE255 pKa = 6.02NGEE258 pKa = 4.37KK259 pKa = 9.73KK260 pKa = 10.59DD261 pKa = 3.57EE262 pKa = 4.94AIYY265 pKa = 10.98SVDD268 pKa = 3.49GKK270 pKa = 10.89RR271 pKa = 11.84GMGDD275 pKa = 3.46YY276 pKa = 10.9YY277 pKa = 11.34VIDD280 pKa = 4.8IIAAGSGSTSADD292 pKa = 2.78QLSFNPSATLYY303 pKa = 7.84WHH305 pKa = 6.52EE306 pKa = 4.2RR307 pKa = 3.47
MM1 pKa = 7.89PYY3 pKa = 10.18LRR5 pKa = 11.84KK6 pKa = 9.48RR7 pKa = 11.84RR8 pKa = 11.84YY9 pKa = 9.05ARR11 pKa = 11.84PRR13 pKa = 11.84YY14 pKa = 9.17SSYY17 pKa = 9.43RR18 pKa = 11.84AKK20 pKa = 10.52RR21 pKa = 11.84RR22 pKa = 11.84TPSSRR27 pKa = 11.84APYY30 pKa = 9.19RR31 pKa = 11.84YY32 pKa = 9.34SRR34 pKa = 11.84KK35 pKa = 8.56RR36 pKa = 11.84TISRR40 pKa = 11.84PRR42 pKa = 11.84YY43 pKa = 8.07AMSRR47 pKa = 11.84KK48 pKa = 9.16RR49 pKa = 11.84LLNITSRR56 pKa = 11.84KK57 pKa = 9.19KK58 pKa = 10.18QDD60 pKa = 3.04TMLVNTNTVAGTPFGGTTFSNTAALLQGGNTYY92 pKa = 10.35IFPWVATARR101 pKa = 11.84DD102 pKa = 3.94NTIGTASGNVFNSADD117 pKa = 3.76RR118 pKa = 11.84TATTCYY124 pKa = 9.66MRR126 pKa = 11.84GLKK129 pKa = 9.81EE130 pKa = 4.23RR131 pKa = 11.84IQIQTNTGMPWQWRR145 pKa = 11.84RR146 pKa = 11.84ICFTMKK152 pKa = 10.67GDD154 pKa = 6.5DD155 pKa = 3.21INAWNEE161 pKa = 3.91SAYY164 pKa = 9.95KK165 pKa = 10.41LQLEE169 pKa = 4.8TSNGWARR176 pKa = 11.84VVSNTGVASNLYY188 pKa = 9.9NNLVPLLFKK197 pKa = 10.7GAQNVDD203 pKa = 2.78WNNPFNAKK211 pKa = 8.72TDD213 pKa = 3.57NTRR216 pKa = 11.84LSIKK220 pKa = 9.94YY221 pKa = 10.31DD222 pKa = 2.75KK223 pKa = 7.68TTPIRR228 pKa = 11.84CGNSNGMMKK237 pKa = 10.28EE238 pKa = 4.27FIRR241 pKa = 11.84WHH243 pKa = 6.23PMNKK247 pKa = 9.58NLVYY251 pKa = 10.83DD252 pKa = 4.27DD253 pKa = 5.28DD254 pKa = 4.83EE255 pKa = 6.02NGEE258 pKa = 4.37KK259 pKa = 9.73KK260 pKa = 10.59DD261 pKa = 3.57EE262 pKa = 4.94AIYY265 pKa = 10.98SVDD268 pKa = 3.49GKK270 pKa = 10.89RR271 pKa = 11.84GMGDD275 pKa = 3.46YY276 pKa = 10.9YY277 pKa = 11.34VIDD280 pKa = 4.8IIAAGSGSTSADD292 pKa = 2.78QLSFNPSATLYY303 pKa = 7.84WHH305 pKa = 6.52EE306 pKa = 4.2RR307 pKa = 3.47
Molecular weight: 35.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
874 |
229 |
338 |
291.3 |
32.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.696 ± 0.493 | 1.259 ± 0.171 |
6.407 ± 0.547 | 4.691 ± 0.858 |
3.547 ± 0.25 | 9.84 ± 1.424 |
1.945 ± 0.539 | 3.776 ± 0.488 |
4.348 ± 0.617 | 7.895 ± 1.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.059 ± 0.517 | 4.691 ± 1.68 |
6.064 ± 1.09 | 2.632 ± 0.015 |
8.009 ± 0.626 | 6.178 ± 0.535 |
5.606 ± 1.445 | 5.606 ± 0.802 |
2.632 ± 0.369 | 4.119 ± 0.58 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
