
Nocardioides sp. JQ2195
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
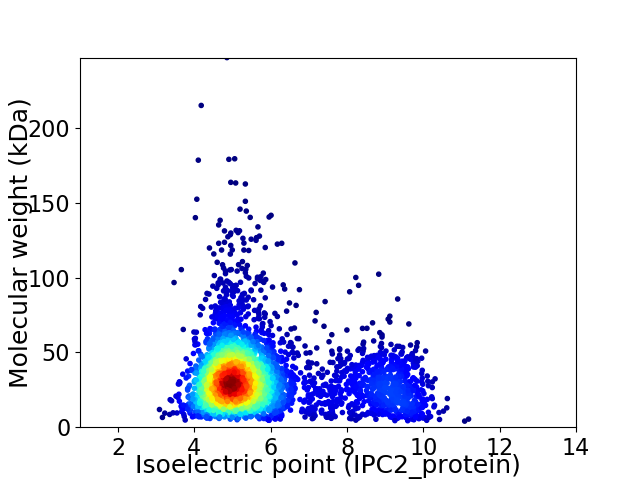
Virtual 2D-PAGE plot for 3770 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
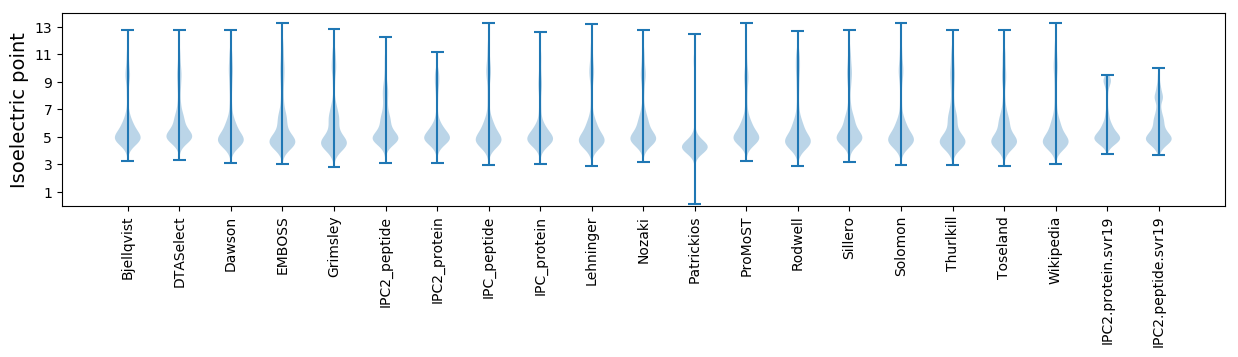
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6H1A056|A0A6H1A056_9ACTN Phospho-sugar mutase OS=Nocardioides sp. JQ2195 OX=2592334 GN=ncot_04225 PE=3 SV=1
MM1 pKa = 7.18HH2 pKa = 6.77SAYY5 pKa = 10.14RR6 pKa = 11.84RR7 pKa = 11.84ASSFRR12 pKa = 11.84QLLARR17 pKa = 11.84MAAVLLISLAAVAGLAGPAAADD39 pKa = 4.5GEE41 pKa = 4.43WSPPDD46 pKa = 3.53GTTSASPSAPADD58 pKa = 3.71DD59 pKa = 5.1AVPAPDD65 pKa = 3.87TPAEE69 pKa = 4.54PEE71 pKa = 4.18TQAPTSAEE79 pKa = 3.95PAAEE83 pKa = 4.03PAEE86 pKa = 4.49EE87 pKa = 4.19PAEE90 pKa = 3.99EE91 pKa = 4.47APTPEE96 pKa = 4.27EE97 pKa = 4.09PKK99 pKa = 10.49AAEE102 pKa = 4.23PKK104 pKa = 10.37AGEE107 pKa = 4.32PADD110 pKa = 3.94EE111 pKa = 4.39QPTPEE116 pKa = 4.66TKK118 pKa = 10.0PAPAKK123 pKa = 9.62STSTTEE129 pKa = 3.49AALAPTTDD137 pKa = 3.73GPEE140 pKa = 4.03SKK142 pKa = 10.58VWICKK147 pKa = 9.9YY148 pKa = 10.83SSTPEE153 pKa = 4.18DD154 pKa = 3.34NEE156 pKa = 3.91KK157 pKa = 10.8AQTVNSVAYY166 pKa = 9.51NSNRR170 pKa = 11.84IPGTWGFEE178 pKa = 4.04DD179 pKa = 4.04AQGLTYY185 pKa = 10.49VYY187 pKa = 10.99SFDD190 pKa = 3.32TGQASKK196 pKa = 10.34PSKK199 pKa = 10.33DD200 pKa = 3.66DD201 pKa = 3.7CPVLIDD207 pKa = 4.48PPAPPTWTEE216 pKa = 3.93PTCADD221 pKa = 3.9PLGSYY226 pKa = 8.3QLPAYY231 pKa = 8.66PADD234 pKa = 4.0RR235 pKa = 11.84ATVSTSGSLEE245 pKa = 3.9PGGTFTVTWTAVSPYY260 pKa = 10.36RR261 pKa = 11.84FLNGSGAPTEE271 pKa = 4.56TISFSHH277 pKa = 6.67TFSQLSSCPVVIDD290 pKa = 4.89DD291 pKa = 4.83LPTVQVSDD299 pKa = 3.74PCGTADD305 pKa = 4.65DD306 pKa = 4.73SVTLSHH312 pKa = 6.84SSQYY316 pKa = 10.76TGVDD320 pKa = 3.3NGDD323 pKa = 3.25GTATFTAVPPNVFPGNEE340 pKa = 3.42ATYY343 pKa = 9.53TVTYY347 pKa = 10.69VPTTDD352 pKa = 3.38VPCIVTVDD360 pKa = 4.11LAAPTITPSTACDD373 pKa = 4.01TPWSIAIGDD382 pKa = 4.0TDD384 pKa = 3.85HH385 pKa = 6.83VSYY388 pKa = 10.6AISPAASGTEE398 pKa = 3.85ATTVTITATPDD409 pKa = 2.9VFFAFSPAPGWTIHH423 pKa = 7.15PDD425 pKa = 2.96GTATFEE431 pKa = 4.3HH432 pKa = 6.73SFAAATTCLDD442 pKa = 3.57VPAEE446 pKa = 4.53PIVTAPTCYY455 pKa = 10.3DD456 pKa = 3.36DD457 pKa = 5.99GYY459 pKa = 9.56LTVEE463 pKa = 4.25IRR465 pKa = 11.84DD466 pKa = 3.79HH467 pKa = 6.21ATVTVNGTTITSATMFGPGSYY488 pKa = 10.48DD489 pKa = 3.25VVYY492 pKa = 9.09TADD495 pKa = 3.64AGHH498 pKa = 6.36VFDD501 pKa = 6.16DD502 pKa = 4.71AGTEE506 pKa = 4.39STTTTYY512 pKa = 11.72DD513 pKa = 4.0DD514 pKa = 3.87IVVEE518 pKa = 4.54AATGDD523 pKa = 4.03CVIDD527 pKa = 4.37VPAQPDD533 pKa = 3.63STDD536 pKa = 2.81TCNPDD541 pKa = 3.27VVVDD545 pKa = 3.93NVEE548 pKa = 3.97WAPFPTLDD556 pKa = 3.94NIVWSVDD563 pKa = 2.83ASTGEE568 pKa = 4.55AIATAINGMSFSNGVTEE585 pKa = 5.05KK586 pKa = 10.47RR587 pKa = 11.84YY588 pKa = 10.45SLPADD593 pKa = 3.51NGIKK597 pKa = 10.09CAAPVDD603 pKa = 4.02PVVTQSSACDD613 pKa = 3.44VQGTLTVPDD622 pKa = 4.09TEE624 pKa = 5.09GITYY628 pKa = 10.69ALDD631 pKa = 4.12GDD633 pKa = 4.73TIAPGTHH640 pKa = 7.26DD641 pKa = 4.43GPLSGTLTATALPGYY656 pKa = 9.81EE657 pKa = 4.56DD658 pKa = 4.42TDD660 pKa = 4.42ADD662 pKa = 3.52WSFHH666 pKa = 5.92VDD668 pKa = 2.81IAAATTCLDD677 pKa = 3.79VPAEE681 pKa = 4.09PAPTPPTCFDD691 pKa = 4.52DD692 pKa = 4.13GTLTVTPAAHH702 pKa = 6.1ATVTVNGDD710 pKa = 3.95PIVDD714 pKa = 3.63EE715 pKa = 4.38TVYY718 pKa = 11.18GPGSYY723 pKa = 10.62DD724 pKa = 3.34VVYY727 pKa = 9.09TADD730 pKa = 3.64AGHH733 pKa = 6.36VFDD736 pKa = 6.16DD737 pKa = 4.71AGTEE741 pKa = 4.39STTTTYY747 pKa = 11.72DD748 pKa = 4.0DD749 pKa = 3.87IVVEE753 pKa = 4.54AATGDD758 pKa = 3.93CVIALEE764 pKa = 4.62AEE766 pKa = 4.06PTEE769 pKa = 4.3IDD771 pKa = 3.8PCNTDD776 pKa = 2.82ATSDD780 pKa = 3.78NIRR783 pKa = 11.84WADD786 pKa = 3.83FPVQDD791 pKa = 5.0DD792 pKa = 4.57IEE794 pKa = 4.4WSVDD798 pKa = 3.12PSTGEE803 pKa = 3.96AVATATNGMFFGPGVTEE820 pKa = 3.74QRR822 pKa = 11.84FAPSPDD828 pKa = 3.47AGLDD832 pKa = 3.55CSLLPGSISSVCVGDD847 pKa = 4.28IPYY850 pKa = 10.55LSYY853 pKa = 11.82AMTLPDD859 pKa = 5.21DD860 pKa = 4.11YY861 pKa = 11.71VGPTDD866 pKa = 3.48MTITFVNPDD875 pKa = 2.74GRR877 pKa = 11.84DD878 pKa = 3.56YY879 pKa = 11.67VVTGQPLAGEE889 pKa = 4.45LLWPGASVDD898 pKa = 3.71PAGWPGWVRR907 pKa = 11.84NPDD910 pKa = 3.09GTYY913 pKa = 10.37SEE915 pKa = 4.61TTGNYY920 pKa = 7.58AWTRR924 pKa = 11.84EE925 pKa = 4.0GVEE928 pKa = 3.77VRR930 pKa = 11.84FQVNPEE936 pKa = 3.78FTTTVAYY943 pKa = 8.65PQATSACAGPAPADD957 pKa = 3.89DD958 pKa = 4.71PGEE961 pKa = 4.35GPEE964 pKa = 4.73EE965 pKa = 4.47GPKK968 pKa = 9.99PQAKK972 pKa = 9.77PSTPDD977 pKa = 3.73AILPNTGAPSGSGMYY992 pKa = 10.21AVLGGLYY999 pKa = 9.12LTFGAWLMFRR1009 pKa = 11.84NRR1011 pKa = 11.84RR1012 pKa = 11.84RR1013 pKa = 11.84RR1014 pKa = 11.84AA1015 pKa = 3.04
MM1 pKa = 7.18HH2 pKa = 6.77SAYY5 pKa = 10.14RR6 pKa = 11.84RR7 pKa = 11.84ASSFRR12 pKa = 11.84QLLARR17 pKa = 11.84MAAVLLISLAAVAGLAGPAAADD39 pKa = 4.5GEE41 pKa = 4.43WSPPDD46 pKa = 3.53GTTSASPSAPADD58 pKa = 3.71DD59 pKa = 5.1AVPAPDD65 pKa = 3.87TPAEE69 pKa = 4.54PEE71 pKa = 4.18TQAPTSAEE79 pKa = 3.95PAAEE83 pKa = 4.03PAEE86 pKa = 4.49EE87 pKa = 4.19PAEE90 pKa = 3.99EE91 pKa = 4.47APTPEE96 pKa = 4.27EE97 pKa = 4.09PKK99 pKa = 10.49AAEE102 pKa = 4.23PKK104 pKa = 10.37AGEE107 pKa = 4.32PADD110 pKa = 3.94EE111 pKa = 4.39QPTPEE116 pKa = 4.66TKK118 pKa = 10.0PAPAKK123 pKa = 9.62STSTTEE129 pKa = 3.49AALAPTTDD137 pKa = 3.73GPEE140 pKa = 4.03SKK142 pKa = 10.58VWICKK147 pKa = 9.9YY148 pKa = 10.83SSTPEE153 pKa = 4.18DD154 pKa = 3.34NEE156 pKa = 3.91KK157 pKa = 10.8AQTVNSVAYY166 pKa = 9.51NSNRR170 pKa = 11.84IPGTWGFEE178 pKa = 4.04DD179 pKa = 4.04AQGLTYY185 pKa = 10.49VYY187 pKa = 10.99SFDD190 pKa = 3.32TGQASKK196 pKa = 10.34PSKK199 pKa = 10.33DD200 pKa = 3.66DD201 pKa = 3.7CPVLIDD207 pKa = 4.48PPAPPTWTEE216 pKa = 3.93PTCADD221 pKa = 3.9PLGSYY226 pKa = 8.3QLPAYY231 pKa = 8.66PADD234 pKa = 4.0RR235 pKa = 11.84ATVSTSGSLEE245 pKa = 3.9PGGTFTVTWTAVSPYY260 pKa = 10.36RR261 pKa = 11.84FLNGSGAPTEE271 pKa = 4.56TISFSHH277 pKa = 6.67TFSQLSSCPVVIDD290 pKa = 4.89DD291 pKa = 4.83LPTVQVSDD299 pKa = 3.74PCGTADD305 pKa = 4.65DD306 pKa = 4.73SVTLSHH312 pKa = 6.84SSQYY316 pKa = 10.76TGVDD320 pKa = 3.3NGDD323 pKa = 3.25GTATFTAVPPNVFPGNEE340 pKa = 3.42ATYY343 pKa = 9.53TVTYY347 pKa = 10.69VPTTDD352 pKa = 3.38VPCIVTVDD360 pKa = 4.11LAAPTITPSTACDD373 pKa = 4.01TPWSIAIGDD382 pKa = 4.0TDD384 pKa = 3.85HH385 pKa = 6.83VSYY388 pKa = 10.6AISPAASGTEE398 pKa = 3.85ATTVTITATPDD409 pKa = 2.9VFFAFSPAPGWTIHH423 pKa = 7.15PDD425 pKa = 2.96GTATFEE431 pKa = 4.3HH432 pKa = 6.73SFAAATTCLDD442 pKa = 3.57VPAEE446 pKa = 4.53PIVTAPTCYY455 pKa = 10.3DD456 pKa = 3.36DD457 pKa = 5.99GYY459 pKa = 9.56LTVEE463 pKa = 4.25IRR465 pKa = 11.84DD466 pKa = 3.79HH467 pKa = 6.21ATVTVNGTTITSATMFGPGSYY488 pKa = 10.48DD489 pKa = 3.25VVYY492 pKa = 9.09TADD495 pKa = 3.64AGHH498 pKa = 6.36VFDD501 pKa = 6.16DD502 pKa = 4.71AGTEE506 pKa = 4.39STTTTYY512 pKa = 11.72DD513 pKa = 4.0DD514 pKa = 3.87IVVEE518 pKa = 4.54AATGDD523 pKa = 4.03CVIDD527 pKa = 4.37VPAQPDD533 pKa = 3.63STDD536 pKa = 2.81TCNPDD541 pKa = 3.27VVVDD545 pKa = 3.93NVEE548 pKa = 3.97WAPFPTLDD556 pKa = 3.94NIVWSVDD563 pKa = 2.83ASTGEE568 pKa = 4.55AIATAINGMSFSNGVTEE585 pKa = 5.05KK586 pKa = 10.47RR587 pKa = 11.84YY588 pKa = 10.45SLPADD593 pKa = 3.51NGIKK597 pKa = 10.09CAAPVDD603 pKa = 4.02PVVTQSSACDD613 pKa = 3.44VQGTLTVPDD622 pKa = 4.09TEE624 pKa = 5.09GITYY628 pKa = 10.69ALDD631 pKa = 4.12GDD633 pKa = 4.73TIAPGTHH640 pKa = 7.26DD641 pKa = 4.43GPLSGTLTATALPGYY656 pKa = 9.81EE657 pKa = 4.56DD658 pKa = 4.42TDD660 pKa = 4.42ADD662 pKa = 3.52WSFHH666 pKa = 5.92VDD668 pKa = 2.81IAAATTCLDD677 pKa = 3.79VPAEE681 pKa = 4.09PAPTPPTCFDD691 pKa = 4.52DD692 pKa = 4.13GTLTVTPAAHH702 pKa = 6.1ATVTVNGDD710 pKa = 3.95PIVDD714 pKa = 3.63EE715 pKa = 4.38TVYY718 pKa = 11.18GPGSYY723 pKa = 10.62DD724 pKa = 3.34VVYY727 pKa = 9.09TADD730 pKa = 3.64AGHH733 pKa = 6.36VFDD736 pKa = 6.16DD737 pKa = 4.71AGTEE741 pKa = 4.39STTTTYY747 pKa = 11.72DD748 pKa = 4.0DD749 pKa = 3.87IVVEE753 pKa = 4.54AATGDD758 pKa = 3.93CVIALEE764 pKa = 4.62AEE766 pKa = 4.06PTEE769 pKa = 4.3IDD771 pKa = 3.8PCNTDD776 pKa = 2.82ATSDD780 pKa = 3.78NIRR783 pKa = 11.84WADD786 pKa = 3.83FPVQDD791 pKa = 5.0DD792 pKa = 4.57IEE794 pKa = 4.4WSVDD798 pKa = 3.12PSTGEE803 pKa = 3.96AVATATNGMFFGPGVTEE820 pKa = 3.74QRR822 pKa = 11.84FAPSPDD828 pKa = 3.47AGLDD832 pKa = 3.55CSLLPGSISSVCVGDD847 pKa = 4.28IPYY850 pKa = 10.55LSYY853 pKa = 11.82AMTLPDD859 pKa = 5.21DD860 pKa = 4.11YY861 pKa = 11.71VGPTDD866 pKa = 3.48MTITFVNPDD875 pKa = 2.74GRR877 pKa = 11.84DD878 pKa = 3.56YY879 pKa = 11.67VVTGQPLAGEE889 pKa = 4.45LLWPGASVDD898 pKa = 3.71PAGWPGWVRR907 pKa = 11.84NPDD910 pKa = 3.09GTYY913 pKa = 10.37SEE915 pKa = 4.61TTGNYY920 pKa = 7.58AWTRR924 pKa = 11.84EE925 pKa = 4.0GVEE928 pKa = 3.77VRR930 pKa = 11.84FQVNPEE936 pKa = 3.78FTTTVAYY943 pKa = 8.65PQATSACAGPAPADD957 pKa = 3.89DD958 pKa = 4.71PGEE961 pKa = 4.35GPEE964 pKa = 4.73EE965 pKa = 4.47GPKK968 pKa = 9.99PQAKK972 pKa = 9.77PSTPDD977 pKa = 3.73AILPNTGAPSGSGMYY992 pKa = 10.21AVLGGLYY999 pKa = 9.12LTFGAWLMFRR1009 pKa = 11.84NRR1011 pKa = 11.84RR1012 pKa = 11.84RR1013 pKa = 11.84RR1014 pKa = 11.84AA1015 pKa = 3.04
Molecular weight: 105.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6H1A5L6|A0A6H1A5L6_9ACTN Helix-turn-helix transcriptional regulator OS=Nocardioides sp. JQ2195 OX=2592334 GN=ncot_17925 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
Molecular weight: 4.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1216581 |
33 |
2276 |
322.7 |
34.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.251 ± 0.048 | 0.75 ± 0.01 |
6.687 ± 0.035 | 6.039 ± 0.038 |
2.96 ± 0.023 | 9.017 ± 0.036 |
2.325 ± 0.02 | 3.83 ± 0.025 |
2.393 ± 0.031 | 9.965 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.026 ± 0.015 | 2.052 ± 0.022 |
5.366 ± 0.028 | 2.849 ± 0.021 |
7.304 ± 0.043 | 5.669 ± 0.029 |
6.044 ± 0.035 | 9.108 ± 0.034 |
1.514 ± 0.016 | 1.852 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
