
Nocardiopsis sp. NRRL B-16309
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Nocardiopsis; unclassified Nocardiopsis
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
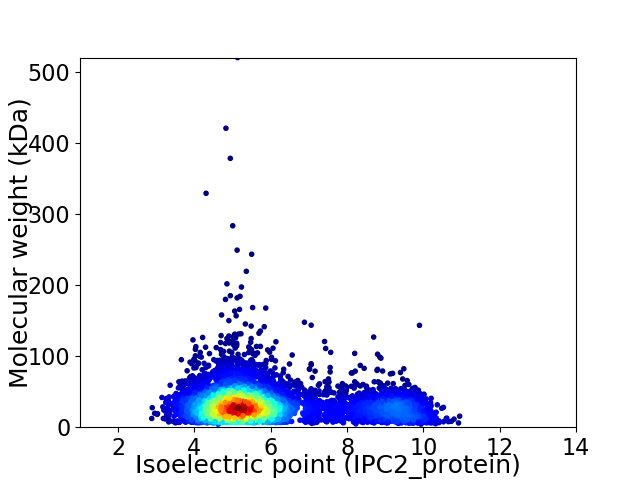
Virtual 2D-PAGE plot for 5253 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N0ARF7|A0A0N0ARF7_9ACTN Glycosyl hydrolase family 26 OS=Nocardiopsis sp. NRRL B-16309 OX=1519494 GN=ADL05_04950 PE=3 SV=1
MM1 pKa = 7.91PPRR4 pKa = 11.84TSPRR8 pKa = 11.84HH9 pKa = 4.82LAAPAAPIRR18 pKa = 11.84SAALTASAALAALTLLGACAADD40 pKa = 4.55PDD42 pKa = 4.47PAPDD46 pKa = 3.39QDD48 pKa = 4.21AGTGADD54 pKa = 4.81FPAASLDD61 pKa = 3.53GLRR64 pKa = 11.84VLLANDD70 pKa = 4.48DD71 pKa = 3.86SMQAGEE77 pKa = 4.48EE78 pKa = 4.22DD79 pKa = 4.05GSDD82 pKa = 3.38GLGLYY87 pKa = 7.51EE88 pKa = 5.76LRR90 pKa = 11.84SALCAAGADD99 pKa = 3.54VVVFAPWGYY108 pKa = 9.44QSSMSSAISHH118 pKa = 6.14SGSFGLGAHH127 pKa = 7.16PGPPEE132 pKa = 4.43EE133 pKa = 4.32YY134 pKa = 10.77AGDD137 pKa = 4.07CADD140 pKa = 4.38APSGGAVHH148 pKa = 6.52GVCVADD154 pKa = 5.79GPCEE158 pKa = 4.31DD159 pKa = 5.47DD160 pKa = 4.68SPSATPVDD168 pKa = 4.03SVTFALHH175 pKa = 6.46HH176 pKa = 6.19GLSEE180 pKa = 4.13LVGWDD185 pKa = 3.87GPPDD189 pKa = 3.64LVVSGVNSGPNVASQIANSGTAGAAFAGQGAGVPAIAVSAGLDD232 pKa = 3.37EE233 pKa = 5.58DD234 pKa = 4.1FTVAPRR240 pKa = 11.84TYY242 pKa = 8.86TAAAEE247 pKa = 4.15FSTDD251 pKa = 2.94LVARR255 pKa = 11.84LVGSDD260 pKa = 4.68LLTSDD265 pKa = 3.51YY266 pKa = 9.89MINVNHH272 pKa = 6.51PHH274 pKa = 6.76APDD277 pKa = 4.04GAPGTDD283 pKa = 3.49VRR285 pKa = 11.84WTRR288 pKa = 11.84AGTGTVLIPEE298 pKa = 4.59FTGQDD303 pKa = 3.28AYY305 pKa = 10.65EE306 pKa = 4.36LGVRR310 pKa = 11.84VCEE313 pKa = 4.51PDD315 pKa = 3.18TPGCVPEE322 pKa = 4.64TSADD326 pKa = 3.73ADD328 pKa = 3.93STALLAEE335 pKa = 4.54GAVSVSALTADD346 pKa = 3.43RR347 pKa = 11.84TYY349 pKa = 11.78ASGEE353 pKa = 3.97DD354 pKa = 3.53PAEE357 pKa = 4.12VEE359 pKa = 4.18RR360 pKa = 11.84LAEE363 pKa = 3.98LVAALGGG370 pKa = 3.68
MM1 pKa = 7.91PPRR4 pKa = 11.84TSPRR8 pKa = 11.84HH9 pKa = 4.82LAAPAAPIRR18 pKa = 11.84SAALTASAALAALTLLGACAADD40 pKa = 4.55PDD42 pKa = 4.47PAPDD46 pKa = 3.39QDD48 pKa = 4.21AGTGADD54 pKa = 4.81FPAASLDD61 pKa = 3.53GLRR64 pKa = 11.84VLLANDD70 pKa = 4.48DD71 pKa = 3.86SMQAGEE77 pKa = 4.48EE78 pKa = 4.22DD79 pKa = 4.05GSDD82 pKa = 3.38GLGLYY87 pKa = 7.51EE88 pKa = 5.76LRR90 pKa = 11.84SALCAAGADD99 pKa = 3.54VVVFAPWGYY108 pKa = 9.44QSSMSSAISHH118 pKa = 6.14SGSFGLGAHH127 pKa = 7.16PGPPEE132 pKa = 4.43EE133 pKa = 4.32YY134 pKa = 10.77AGDD137 pKa = 4.07CADD140 pKa = 4.38APSGGAVHH148 pKa = 6.52GVCVADD154 pKa = 5.79GPCEE158 pKa = 4.31DD159 pKa = 5.47DD160 pKa = 4.68SPSATPVDD168 pKa = 4.03SVTFALHH175 pKa = 6.46HH176 pKa = 6.19GLSEE180 pKa = 4.13LVGWDD185 pKa = 3.87GPPDD189 pKa = 3.64LVVSGVNSGPNVASQIANSGTAGAAFAGQGAGVPAIAVSAGLDD232 pKa = 3.37EE233 pKa = 5.58DD234 pKa = 4.1FTVAPRR240 pKa = 11.84TYY242 pKa = 8.86TAAAEE247 pKa = 4.15FSTDD251 pKa = 2.94LVARR255 pKa = 11.84LVGSDD260 pKa = 4.68LLTSDD265 pKa = 3.51YY266 pKa = 9.89MINVNHH272 pKa = 6.51PHH274 pKa = 6.76APDD277 pKa = 4.04GAPGTDD283 pKa = 3.49VRR285 pKa = 11.84WTRR288 pKa = 11.84AGTGTVLIPEE298 pKa = 4.59FTGQDD303 pKa = 3.28AYY305 pKa = 10.65EE306 pKa = 4.36LGVRR310 pKa = 11.84VCEE313 pKa = 4.51PDD315 pKa = 3.18TPGCVPEE322 pKa = 4.64TSADD326 pKa = 3.73ADD328 pKa = 3.93STALLAEE335 pKa = 4.54GAVSVSALTADD346 pKa = 3.43RR347 pKa = 11.84TYY349 pKa = 11.78ASGEE353 pKa = 3.97DD354 pKa = 3.53PAEE357 pKa = 4.12VEE359 pKa = 4.18RR360 pKa = 11.84LAEE363 pKa = 3.98LVAALGGG370 pKa = 3.68
Molecular weight: 36.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N0ANB3|A0A0N0ANB3_9ACTN NADH-quinone oxidoreductase subunit I OS=Nocardiopsis sp. NRRL B-16309 OX=1519494 GN=nuoI PE=3 SV=1
MM1 pKa = 7.57LGFLGRR7 pKa = 11.84LRR9 pKa = 11.84LLGRR13 pKa = 11.84LGRR16 pKa = 11.84LPLSHH21 pKa = 7.74PGRR24 pKa = 11.84LGRR27 pKa = 11.84LRR29 pKa = 11.84LVGSLGHH36 pKa = 6.97LPLGPLEE43 pKa = 4.13LRR45 pKa = 11.84GFLRR49 pKa = 11.84LLGLDD54 pKa = 3.09PVMRR58 pKa = 11.84RR59 pKa = 11.84PRR61 pKa = 11.84HH62 pKa = 5.41LGSMGMPSRR71 pKa = 11.84LLHH74 pKa = 6.1GRR76 pKa = 11.84LGTLRR81 pKa = 11.84RR82 pKa = 11.84LLLHH86 pKa = 5.61SLGRR90 pKa = 11.84LGHH93 pKa = 6.51LRR95 pKa = 11.84PIRR98 pKa = 11.84LLVWLLLGRR107 pKa = 11.84LGGLGGAGPVGLPRR121 pKa = 11.84LLDD124 pKa = 3.87LLRR127 pKa = 11.84LRR129 pKa = 11.84ALGLGSVMRR138 pKa = 11.84RR139 pKa = 11.84LGGG142 pKa = 3.31
MM1 pKa = 7.57LGFLGRR7 pKa = 11.84LRR9 pKa = 11.84LLGRR13 pKa = 11.84LGRR16 pKa = 11.84LPLSHH21 pKa = 7.74PGRR24 pKa = 11.84LGRR27 pKa = 11.84LRR29 pKa = 11.84LVGSLGHH36 pKa = 6.97LPLGPLEE43 pKa = 4.13LRR45 pKa = 11.84GFLRR49 pKa = 11.84LLGLDD54 pKa = 3.09PVMRR58 pKa = 11.84RR59 pKa = 11.84PRR61 pKa = 11.84HH62 pKa = 5.41LGSMGMPSRR71 pKa = 11.84LLHH74 pKa = 6.1GRR76 pKa = 11.84LGTLRR81 pKa = 11.84RR82 pKa = 11.84LLLHH86 pKa = 5.61SLGRR90 pKa = 11.84LGHH93 pKa = 6.51LRR95 pKa = 11.84PIRR98 pKa = 11.84LLVWLLLGRR107 pKa = 11.84LGGLGGAGPVGLPRR121 pKa = 11.84LLDD124 pKa = 3.87LLRR127 pKa = 11.84LRR129 pKa = 11.84ALGLGSVMRR138 pKa = 11.84RR139 pKa = 11.84LGGG142 pKa = 3.31
Molecular weight: 15.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1660638 |
37 |
4719 |
316.1 |
33.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.173 ± 0.046 | 0.726 ± 0.009 |
6.506 ± 0.033 | 6.391 ± 0.034 |
2.76 ± 0.019 | 9.464 ± 0.033 |
2.367 ± 0.015 | 3.106 ± 0.023 |
1.469 ± 0.021 | 10.215 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.88 ± 0.014 | 1.662 ± 0.018 |
6.058 ± 0.038 | 2.483 ± 0.019 |
8.29 ± 0.038 | 5.13 ± 0.026 |
5.902 ± 0.028 | 8.927 ± 0.041 |
1.523 ± 0.013 | 1.969 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
