
Ornithinimicrobium sp. CNJ-824
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Ornithinimicrobiaceae; Ornithinimicrobium; unclassified Ornithinimicrobium
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
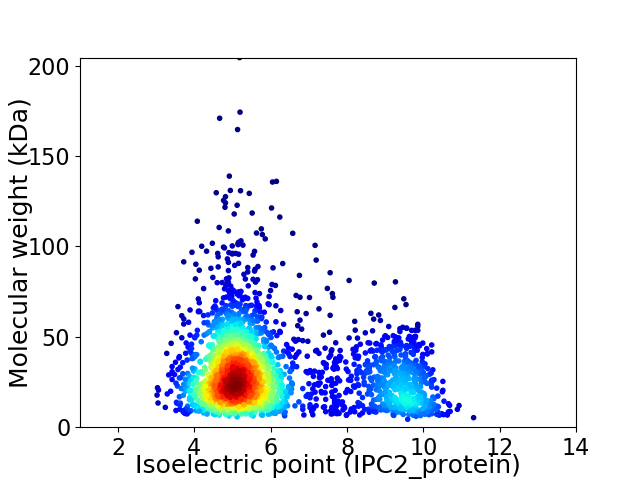
Virtual 2D-PAGE plot for 2650 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
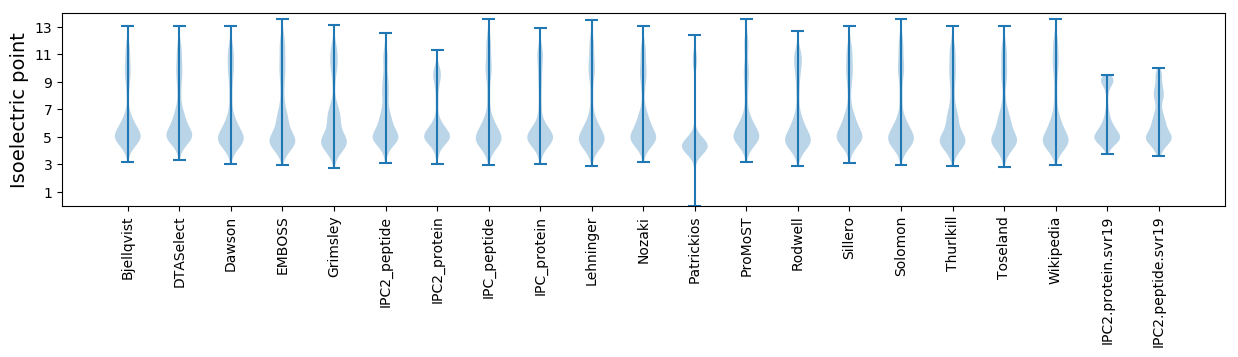
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U7MS04|A0A1U7MS04_9MICO Sugar ABC transporter substrate-binding protein OS=Ornithinimicrobium sp. CNJ-824 OX=1904966 GN=BJF81_13485 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 9.78STGRR6 pKa = 11.84GAFVASLAALALGLSACGSDD26 pKa = 3.41NGDD29 pKa = 3.4GGGGGDD35 pKa = 3.49AGGDD39 pKa = 3.15SGEE42 pKa = 4.29PIKK45 pKa = 11.16VGMIADD51 pKa = 4.16LTGATGDD58 pKa = 3.17VGAPYY63 pKa = 10.58NEE65 pKa = 3.83GMLAYY70 pKa = 10.42VDD72 pKa = 3.55WRR74 pKa = 11.84NEE76 pKa = 3.82NGGVADD82 pKa = 4.93RR83 pKa = 11.84EE84 pKa = 4.16IEE86 pKa = 4.81AISNDD91 pKa = 3.22YY92 pKa = 10.8AYY94 pKa = 9.43EE95 pKa = 4.23VPQAEE100 pKa = 4.38QLYY103 pKa = 9.7KK104 pKa = 10.59QYY106 pKa = 11.4LNDD109 pKa = 3.44GVVAVQGWGTGDD121 pKa = 3.33TEE123 pKa = 4.26ALHH126 pKa = 6.23TKK128 pKa = 10.0VADD131 pKa = 3.91DD132 pKa = 4.63EE133 pKa = 4.8LPFMSGSFAEE143 pKa = 4.45SLTDD147 pKa = 4.28PEE149 pKa = 3.97EE150 pKa = 4.24APYY153 pKa = 11.02NFVVAPTYY161 pKa = 10.37SDD163 pKa = 3.4QMRR166 pKa = 11.84VALNWIAEE174 pKa = 4.13DD175 pKa = 3.47SGGGAEE181 pKa = 4.14VAVFHH186 pKa = 7.1HH187 pKa = 7.22DD188 pKa = 3.63SPFGQAPVADD198 pKa = 4.06GEE200 pKa = 4.48AWVSEE205 pKa = 4.11EE206 pKa = 4.49GLDD209 pKa = 5.07LGYY212 pKa = 10.16QAYY215 pKa = 10.4AMPGGQQNYY224 pKa = 10.16VGLLSQAQSQGAEE237 pKa = 3.95YY238 pKa = 10.32IVIQNVASPAAQVAKK253 pKa = 10.65DD254 pKa = 3.33IAAQNLDD261 pKa = 3.57MKK263 pKa = 10.33IVCLNWCGNEE273 pKa = 4.08LFIDD277 pKa = 3.69TAGADD282 pKa = 3.56VAEE285 pKa = 4.38GHH287 pKa = 6.41MLVQPFAPITSEE299 pKa = 4.16KK300 pKa = 10.45AGHH303 pKa = 6.16EE304 pKa = 4.1EE305 pKa = 3.45MTAYY309 pKa = 10.7LEE311 pKa = 4.3EE312 pKa = 4.29QGEE315 pKa = 4.44DD316 pKa = 3.46PASKK320 pKa = 8.92GTSWVQGWMVMHH332 pKa = 6.26VMAEE336 pKa = 4.11GMEE339 pKa = 4.19KK340 pKa = 10.26AAEE343 pKa = 4.55DD344 pKa = 4.42GEE346 pKa = 4.58ITGPAIRR353 pKa = 11.84EE354 pKa = 4.02ALEE357 pKa = 3.9TMGAIDD363 pKa = 3.5TGGVIGEE370 pKa = 4.35GNIEE374 pKa = 4.31FSADD378 pKa = 3.05SHH380 pKa = 6.85RR381 pKa = 11.84GSTSTGIYY389 pKa = 7.65TAEE392 pKa = 4.23GGEE395 pKa = 4.18MVEE398 pKa = 4.54VEE400 pKa = 4.95AGATPP405 pKa = 3.93
MM1 pKa = 7.63KK2 pKa = 9.78STGRR6 pKa = 11.84GAFVASLAALALGLSACGSDD26 pKa = 3.41NGDD29 pKa = 3.4GGGGGDD35 pKa = 3.49AGGDD39 pKa = 3.15SGEE42 pKa = 4.29PIKK45 pKa = 11.16VGMIADD51 pKa = 4.16LTGATGDD58 pKa = 3.17VGAPYY63 pKa = 10.58NEE65 pKa = 3.83GMLAYY70 pKa = 10.42VDD72 pKa = 3.55WRR74 pKa = 11.84NEE76 pKa = 3.82NGGVADD82 pKa = 4.93RR83 pKa = 11.84EE84 pKa = 4.16IEE86 pKa = 4.81AISNDD91 pKa = 3.22YY92 pKa = 10.8AYY94 pKa = 9.43EE95 pKa = 4.23VPQAEE100 pKa = 4.38QLYY103 pKa = 9.7KK104 pKa = 10.59QYY106 pKa = 11.4LNDD109 pKa = 3.44GVVAVQGWGTGDD121 pKa = 3.33TEE123 pKa = 4.26ALHH126 pKa = 6.23TKK128 pKa = 10.0VADD131 pKa = 3.91DD132 pKa = 4.63EE133 pKa = 4.8LPFMSGSFAEE143 pKa = 4.45SLTDD147 pKa = 4.28PEE149 pKa = 3.97EE150 pKa = 4.24APYY153 pKa = 11.02NFVVAPTYY161 pKa = 10.37SDD163 pKa = 3.4QMRR166 pKa = 11.84VALNWIAEE174 pKa = 4.13DD175 pKa = 3.47SGGGAEE181 pKa = 4.14VAVFHH186 pKa = 7.1HH187 pKa = 7.22DD188 pKa = 3.63SPFGQAPVADD198 pKa = 4.06GEE200 pKa = 4.48AWVSEE205 pKa = 4.11EE206 pKa = 4.49GLDD209 pKa = 5.07LGYY212 pKa = 10.16QAYY215 pKa = 10.4AMPGGQQNYY224 pKa = 10.16VGLLSQAQSQGAEE237 pKa = 3.95YY238 pKa = 10.32IVIQNVASPAAQVAKK253 pKa = 10.65DD254 pKa = 3.33IAAQNLDD261 pKa = 3.57MKK263 pKa = 10.33IVCLNWCGNEE273 pKa = 4.08LFIDD277 pKa = 3.69TAGADD282 pKa = 3.56VAEE285 pKa = 4.38GHH287 pKa = 6.41MLVQPFAPITSEE299 pKa = 4.16KK300 pKa = 10.45AGHH303 pKa = 6.16EE304 pKa = 4.1EE305 pKa = 3.45MTAYY309 pKa = 10.7LEE311 pKa = 4.3EE312 pKa = 4.29QGEE315 pKa = 4.44DD316 pKa = 3.46PASKK320 pKa = 8.92GTSWVQGWMVMHH332 pKa = 6.26VMAEE336 pKa = 4.11GMEE339 pKa = 4.19KK340 pKa = 10.26AAEE343 pKa = 4.55DD344 pKa = 4.42GEE346 pKa = 4.58ITGPAIRR353 pKa = 11.84EE354 pKa = 4.02ALEE357 pKa = 3.9TMGAIDD363 pKa = 3.5TGGVIGEE370 pKa = 4.35GNIEE374 pKa = 4.31FSADD378 pKa = 3.05SHH380 pKa = 6.85RR381 pKa = 11.84GSTSTGIYY389 pKa = 7.65TAEE392 pKa = 4.23GGEE395 pKa = 4.18MVEE398 pKa = 4.54VEE400 pKa = 4.95AGATPP405 pKa = 3.93
Molecular weight: 42.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U7MN07|A0A1U7MN07_9MICO Uncharacterized protein OS=Ornithinimicrobium sp. CNJ-824 OX=1904966 GN=BJF81_07780 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84KK16 pKa = 8.65HH17 pKa = 4.47GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.41GRR40 pKa = 11.84SSLSAA45 pKa = 3.62
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84KK16 pKa = 8.65HH17 pKa = 4.47GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.41GRR40 pKa = 11.84SSLSAA45 pKa = 3.62
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
792233 |
37 |
1914 |
299.0 |
31.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.989 ± 0.063 | 0.658 ± 0.015 |
6.403 ± 0.044 | 6.071 ± 0.046 |
2.408 ± 0.028 | 9.681 ± 0.052 |
2.321 ± 0.026 | 2.657 ± 0.031 |
1.362 ± 0.028 | 10.612 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.888 ± 0.021 | 1.359 ± 0.021 |
6.033 ± 0.04 | 2.944 ± 0.026 |
8.321 ± 0.048 | 4.75 ± 0.03 |
6.08 ± 0.039 | 10.084 ± 0.05 |
1.654 ± 0.022 | 1.723 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
