
Plasmodium falciparum UGT5.1
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Laverania); Plasmodium falciparum
Average proteome isoelectric point is 7.25
Get precalculated fractions of proteins
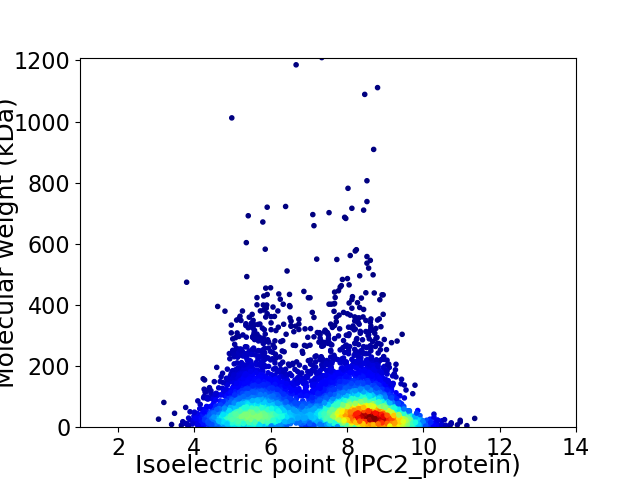
Virtual 2D-PAGE plot for 5904 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
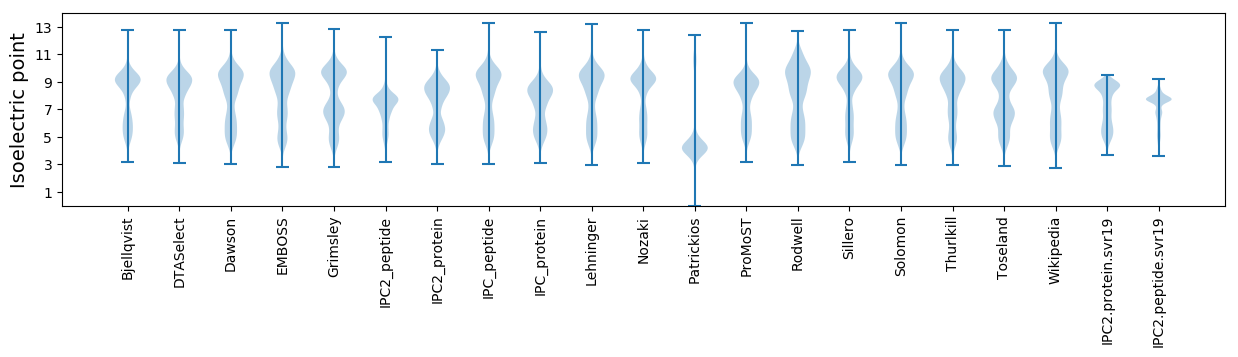
Protein with the lowest isoelectric point:
>tr|W7JYT6|W7JYT6_PLAFA Uncharacterized protein OS=Plasmodium falciparum UGT5.1 OX=1237627 GN=C923_02547 PE=4 SV=1
MM1 pKa = 7.58EE2 pKa = 6.01EE3 pKa = 3.93IVDD6 pKa = 4.39DD7 pKa = 4.22GMEE10 pKa = 4.32EE11 pKa = 3.7IDD13 pKa = 4.67LNEE16 pKa = 4.22NSDD19 pKa = 4.44FIHH22 pKa = 7.4DD23 pKa = 4.13GNPLGEE29 pKa = 4.28YY30 pKa = 10.96SNMEE34 pKa = 4.0YY35 pKa = 10.9EE36 pKa = 4.48LLNEE40 pKa = 4.48VSASTDD46 pKa = 3.38FSDD49 pKa = 3.5YY50 pKa = 11.01HH51 pKa = 7.75SVVDD55 pKa = 3.89EE56 pKa = 4.68DD57 pKa = 5.63LINFNDD63 pKa = 3.99DD64 pKa = 3.11VTLEE68 pKa = 4.2TQSMIAHH75 pKa = 6.92GGSLSEE81 pKa = 4.27IEE83 pKa = 4.17EE84 pKa = 4.39TGDD87 pKa = 3.66LSSDD91 pKa = 3.24VDD93 pKa = 3.74RR94 pKa = 11.84LLSSIEE100 pKa = 3.84TTPRR104 pKa = 11.84DD105 pKa = 3.48DD106 pKa = 3.26VVYY109 pKa = 10.56IRR111 pKa = 11.84GDD113 pKa = 3.33IILGEE118 pKa = 4.14DD119 pKa = 3.84EE120 pKa = 5.19IIIEE124 pKa = 4.18NGEE127 pKa = 4.12IILNNTDD134 pKa = 3.64EE135 pKa = 4.7LFEE138 pKa = 5.13DD139 pKa = 4.03VPEE142 pKa = 4.08MLEE145 pKa = 4.01NANLLEE151 pKa = 4.49DD152 pKa = 3.8RR153 pKa = 11.84EE154 pKa = 4.66HH155 pKa = 7.44VFEE158 pKa = 4.42YY159 pKa = 10.54NISVANEE166 pKa = 3.48NTTNQNSYY174 pKa = 10.71NFNAPQDD181 pKa = 4.02NTLTNYY187 pKa = 10.32NISRR191 pKa = 11.84FLQRR195 pKa = 11.84YY196 pKa = 6.69GTTTPNTQHH205 pKa = 6.75NISIHH210 pKa = 6.53RR211 pKa = 11.84DD212 pKa = 3.18GHH214 pKa = 5.2SLRR217 pKa = 11.84SDD219 pKa = 3.19TNIYY223 pKa = 10.86DD224 pKa = 4.61RR225 pKa = 11.84INGPSIRR232 pKa = 11.84EE233 pKa = 3.67NSLYY237 pKa = 10.28PGSFEE242 pKa = 3.77MAQYY246 pKa = 11.03ADD248 pKa = 4.72RR249 pKa = 11.84YY250 pKa = 10.15QMGHH254 pKa = 6.64TIAEE258 pKa = 4.43GFLYY262 pKa = 10.73
MM1 pKa = 7.58EE2 pKa = 6.01EE3 pKa = 3.93IVDD6 pKa = 4.39DD7 pKa = 4.22GMEE10 pKa = 4.32EE11 pKa = 3.7IDD13 pKa = 4.67LNEE16 pKa = 4.22NSDD19 pKa = 4.44FIHH22 pKa = 7.4DD23 pKa = 4.13GNPLGEE29 pKa = 4.28YY30 pKa = 10.96SNMEE34 pKa = 4.0YY35 pKa = 10.9EE36 pKa = 4.48LLNEE40 pKa = 4.48VSASTDD46 pKa = 3.38FSDD49 pKa = 3.5YY50 pKa = 11.01HH51 pKa = 7.75SVVDD55 pKa = 3.89EE56 pKa = 4.68DD57 pKa = 5.63LINFNDD63 pKa = 3.99DD64 pKa = 3.11VTLEE68 pKa = 4.2TQSMIAHH75 pKa = 6.92GGSLSEE81 pKa = 4.27IEE83 pKa = 4.17EE84 pKa = 4.39TGDD87 pKa = 3.66LSSDD91 pKa = 3.24VDD93 pKa = 3.74RR94 pKa = 11.84LLSSIEE100 pKa = 3.84TTPRR104 pKa = 11.84DD105 pKa = 3.48DD106 pKa = 3.26VVYY109 pKa = 10.56IRR111 pKa = 11.84GDD113 pKa = 3.33IILGEE118 pKa = 4.14DD119 pKa = 3.84EE120 pKa = 5.19IIIEE124 pKa = 4.18NGEE127 pKa = 4.12IILNNTDD134 pKa = 3.64EE135 pKa = 4.7LFEE138 pKa = 5.13DD139 pKa = 4.03VPEE142 pKa = 4.08MLEE145 pKa = 4.01NANLLEE151 pKa = 4.49DD152 pKa = 3.8RR153 pKa = 11.84EE154 pKa = 4.66HH155 pKa = 7.44VFEE158 pKa = 4.42YY159 pKa = 10.54NISVANEE166 pKa = 3.48NTTNQNSYY174 pKa = 10.71NFNAPQDD181 pKa = 4.02NTLTNYY187 pKa = 10.32NISRR191 pKa = 11.84FLQRR195 pKa = 11.84YY196 pKa = 6.69GTTTPNTQHH205 pKa = 6.75NISIHH210 pKa = 6.53RR211 pKa = 11.84DD212 pKa = 3.18GHH214 pKa = 5.2SLRR217 pKa = 11.84SDD219 pKa = 3.19TNIYY223 pKa = 10.86DD224 pKa = 4.61RR225 pKa = 11.84INGPSIRR232 pKa = 11.84EE233 pKa = 3.67NSLYY237 pKa = 10.28PGSFEE242 pKa = 3.77MAQYY246 pKa = 11.03ADD248 pKa = 4.72RR249 pKa = 11.84YY250 pKa = 10.15QMGHH254 pKa = 6.64TIAEE258 pKa = 4.43GFLYY262 pKa = 10.73
Molecular weight: 29.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W7K5D7|W7K5D7_PLAFA Uncharacterized protein OS=Plasmodium falciparum UGT5.1 OX=1237627 GN=C923_00101 PE=3 SV=1
MM1 pKa = 7.39LVNLGPMLVKK11 pKa = 10.76LGPKK15 pKa = 9.81LVIIRR20 pKa = 11.84PISVNLRR27 pKa = 11.84PKK29 pKa = 10.07LVKK32 pKa = 10.38LGPMLVKK39 pKa = 10.39LRR41 pKa = 11.84PMLAKK46 pKa = 10.42LRR48 pKa = 11.84PKK50 pKa = 10.44LVMLGPIFAKK60 pKa = 10.35LRR62 pKa = 11.84PTLVTLRR69 pKa = 11.84PKK71 pKa = 10.75LVMLGPKK78 pKa = 9.91LVTLRR83 pKa = 11.84PTLVTLRR90 pKa = 11.84PTLVTLRR97 pKa = 11.84PTLVTLRR104 pKa = 11.84PKK106 pKa = 10.74YY107 pKa = 11.22VMLGPILLTYY117 pKa = 10.45RR118 pKa = 11.84PILVLVKK125 pKa = 10.34LRR127 pKa = 11.84PVLVKK132 pKa = 10.08VRR134 pKa = 11.84PISVNVRR141 pKa = 11.84PKK143 pKa = 10.83LLVKK147 pKa = 10.55LRR149 pKa = 11.84PKK151 pKa = 9.91LVKK154 pKa = 10.11VRR156 pKa = 11.84PILLKK161 pKa = 10.47VRR163 pKa = 11.84PISVKK168 pKa = 10.1IKK170 pKa = 10.15PKK172 pKa = 10.1SKK174 pKa = 9.77LVKK177 pKa = 9.69VRR179 pKa = 11.84PMLSKK184 pKa = 10.64LRR186 pKa = 11.84SKK188 pKa = 10.28LVKK191 pKa = 10.12VRR193 pKa = 11.84PKK195 pKa = 10.24LVKK198 pKa = 10.06VRR200 pKa = 11.84PKK202 pKa = 10.79LLVTLRR208 pKa = 11.84PMLVKK213 pKa = 10.57LRR215 pKa = 11.84AKK217 pKa = 9.86LVKK220 pKa = 9.84LRR222 pKa = 11.84PMLVKK227 pKa = 10.28VRR229 pKa = 11.84PYY231 pKa = 10.27LVKK234 pKa = 10.61LRR236 pKa = 11.84PLLVNVRR243 pKa = 11.84HH244 pKa = 6.28KK245 pKa = 10.58LVKK248 pKa = 10.13VRR250 pKa = 11.84PP251 pKa = 3.81
MM1 pKa = 7.39LVNLGPMLVKK11 pKa = 10.76LGPKK15 pKa = 9.81LVIIRR20 pKa = 11.84PISVNLRR27 pKa = 11.84PKK29 pKa = 10.07LVKK32 pKa = 10.38LGPMLVKK39 pKa = 10.39LRR41 pKa = 11.84PMLAKK46 pKa = 10.42LRR48 pKa = 11.84PKK50 pKa = 10.44LVMLGPIFAKK60 pKa = 10.35LRR62 pKa = 11.84PTLVTLRR69 pKa = 11.84PKK71 pKa = 10.75LVMLGPKK78 pKa = 9.91LVTLRR83 pKa = 11.84PTLVTLRR90 pKa = 11.84PTLVTLRR97 pKa = 11.84PTLVTLRR104 pKa = 11.84PKK106 pKa = 10.74YY107 pKa = 11.22VMLGPILLTYY117 pKa = 10.45RR118 pKa = 11.84PILVLVKK125 pKa = 10.34LRR127 pKa = 11.84PVLVKK132 pKa = 10.08VRR134 pKa = 11.84PISVNVRR141 pKa = 11.84PKK143 pKa = 10.83LLVKK147 pKa = 10.55LRR149 pKa = 11.84PKK151 pKa = 9.91LVKK154 pKa = 10.11VRR156 pKa = 11.84PILLKK161 pKa = 10.47VRR163 pKa = 11.84PISVKK168 pKa = 10.1IKK170 pKa = 10.15PKK172 pKa = 10.1SKK174 pKa = 9.77LVKK177 pKa = 9.69VRR179 pKa = 11.84PMLSKK184 pKa = 10.64LRR186 pKa = 11.84SKK188 pKa = 10.28LVKK191 pKa = 10.12VRR193 pKa = 11.84PKK195 pKa = 10.24LVKK198 pKa = 10.06VRR200 pKa = 11.84PKK202 pKa = 10.79LLVTLRR208 pKa = 11.84PMLVKK213 pKa = 10.57LRR215 pKa = 11.84AKK217 pKa = 9.86LVKK220 pKa = 9.84LRR222 pKa = 11.84PMLVKK227 pKa = 10.28VRR229 pKa = 11.84PYY231 pKa = 10.27LVKK234 pKa = 10.61LRR236 pKa = 11.84PLLVNVRR243 pKa = 11.84HH244 pKa = 6.28KK245 pKa = 10.58LVKK248 pKa = 10.13VRR250 pKa = 11.84PP251 pKa = 3.81
Molecular weight: 28.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3939455 |
12 |
10314 |
667.3 |
78.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.96 ± 0.024 | 1.798 ± 0.018 |
6.388 ± 0.028 | 6.979 ± 0.039 |
4.399 ± 0.024 | 2.82 ± 0.031 |
2.427 ± 0.013 | 9.308 ± 0.039 |
11.738 ± 0.037 | 7.622 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.212 ± 0.015 | 14.436 ± 0.095 |
1.957 ± 0.02 | 2.766 ± 0.014 |
2.652 ± 0.017 | 6.404 ± 0.024 |
4.11 ± 0.018 | 3.759 ± 0.023 |
0.502 ± 0.009 | 5.763 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
