
Tubeweb spider associated circular virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykolovirus; Gemykolovirus segpa1
Average proteome isoelectric point is 7.51
Get precalculated fractions of proteins
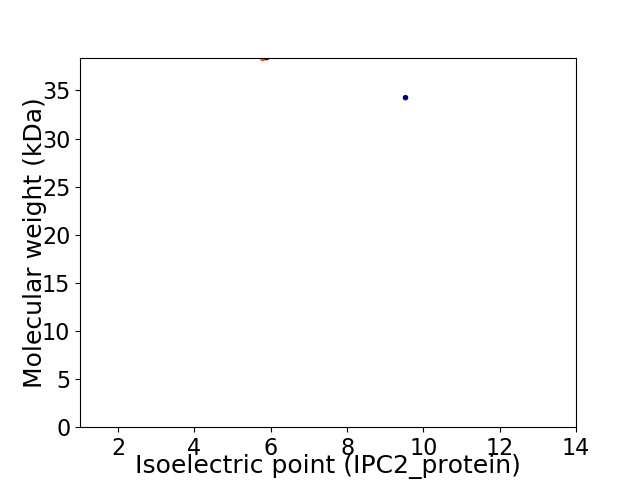
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BP67|A0A346BP67_9VIRU Putative capsid protein OS=Tubeweb spider associated circular virus 1 OX=2293308 PE=4 SV=1
MM1 pKa = 6.51TRR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 10.35KK6 pKa = 10.62LDD8 pKa = 3.18NVEE11 pKa = 4.9YY12 pKa = 10.44ILLTYY17 pKa = 10.29SDD19 pKa = 5.02CPVEE23 pKa = 4.9FDD25 pKa = 3.46PQGIIVAVVRR35 pKa = 11.84SGGVYY40 pKa = 9.4RR41 pKa = 11.84LGRR44 pKa = 11.84EE45 pKa = 3.8LHH47 pKa = 5.79QNGMPHH53 pKa = 5.19YY54 pKa = 9.74HH55 pKa = 6.46CFVWWVGGFSHH66 pKa = 7.9PDD68 pKa = 2.94AGQLFMVSGRR78 pKa = 11.84RR79 pKa = 11.84CNIKK83 pKa = 10.43KK84 pKa = 8.56FTANPGRR91 pKa = 11.84RR92 pKa = 11.84WDD94 pKa = 3.79YY95 pKa = 8.01VGKK98 pKa = 10.48YY99 pKa = 9.56AGHH102 pKa = 7.07KK103 pKa = 9.76DD104 pKa = 3.16GHH106 pKa = 5.64YY107 pKa = 10.66LIGDD111 pKa = 3.37QCEE114 pKa = 4.13RR115 pKa = 11.84PGGDD119 pKa = 3.31KK120 pKa = 10.7DD121 pKa = 4.18DD122 pKa = 4.47SEE124 pKa = 4.46RR125 pKa = 11.84TQTDD129 pKa = 2.27IWSEE133 pKa = 3.96IINATNEE140 pKa = 4.03GEE142 pKa = 4.33FFEE145 pKa = 5.14KK146 pKa = 10.5LAALAPKK153 pKa = 10.22QLGCNFGSLKK163 pKa = 10.72LYY165 pKa = 10.76ADD167 pKa = 3.52WKK169 pKa = 9.79YY170 pKa = 10.4RR171 pKa = 11.84PEE173 pKa = 4.03VADD176 pKa = 3.67YY177 pKa = 8.97EE178 pKa = 4.6SPPGIFEE185 pKa = 4.11PVYY188 pKa = 11.09DD189 pKa = 3.77LTNWSEE195 pKa = 4.07ANLGRR200 pKa = 11.84VEE202 pKa = 3.92GRR204 pKa = 11.84PRR206 pKa = 11.84GLVLFGATRR215 pKa = 11.84LGKK218 pKa = 7.65TVWARR223 pKa = 11.84SLGKK227 pKa = 9.74HH228 pKa = 5.68NYY230 pKa = 8.93FAGLFNLDD238 pKa = 4.56DD239 pKa = 4.25FSEE242 pKa = 4.36SAEE245 pKa = 3.92YY246 pKa = 10.42AIFDD250 pKa = 4.19DD251 pKa = 4.0MSGGFSFFPSYY262 pKa = 9.2KK263 pKa = 9.6QWMGGQFQFTVTDD276 pKa = 3.65KK277 pKa = 10.77FKK279 pKa = 10.89HH280 pKa = 5.41KK281 pKa = 10.03RR282 pKa = 11.84TVRR285 pKa = 11.84WGKK288 pKa = 7.37PTIWLCNTDD297 pKa = 5.47PRR299 pKa = 11.84LDD301 pKa = 3.34WYY303 pKa = 10.98KK304 pKa = 10.58PGSSPDD310 pKa = 3.78FGWMEE315 pKa = 3.92EE316 pKa = 4.02NADD319 pKa = 3.77FVEE322 pKa = 4.27LTEE325 pKa = 4.52TIFRR329 pKa = 11.84ANRR332 pKa = 11.84EE333 pKa = 3.96
MM1 pKa = 6.51TRR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 10.35KK6 pKa = 10.62LDD8 pKa = 3.18NVEE11 pKa = 4.9YY12 pKa = 10.44ILLTYY17 pKa = 10.29SDD19 pKa = 5.02CPVEE23 pKa = 4.9FDD25 pKa = 3.46PQGIIVAVVRR35 pKa = 11.84SGGVYY40 pKa = 9.4RR41 pKa = 11.84LGRR44 pKa = 11.84EE45 pKa = 3.8LHH47 pKa = 5.79QNGMPHH53 pKa = 5.19YY54 pKa = 9.74HH55 pKa = 6.46CFVWWVGGFSHH66 pKa = 7.9PDD68 pKa = 2.94AGQLFMVSGRR78 pKa = 11.84RR79 pKa = 11.84CNIKK83 pKa = 10.43KK84 pKa = 8.56FTANPGRR91 pKa = 11.84RR92 pKa = 11.84WDD94 pKa = 3.79YY95 pKa = 8.01VGKK98 pKa = 10.48YY99 pKa = 9.56AGHH102 pKa = 7.07KK103 pKa = 9.76DD104 pKa = 3.16GHH106 pKa = 5.64YY107 pKa = 10.66LIGDD111 pKa = 3.37QCEE114 pKa = 4.13RR115 pKa = 11.84PGGDD119 pKa = 3.31KK120 pKa = 10.7DD121 pKa = 4.18DD122 pKa = 4.47SEE124 pKa = 4.46RR125 pKa = 11.84TQTDD129 pKa = 2.27IWSEE133 pKa = 3.96IINATNEE140 pKa = 4.03GEE142 pKa = 4.33FFEE145 pKa = 5.14KK146 pKa = 10.5LAALAPKK153 pKa = 10.22QLGCNFGSLKK163 pKa = 10.72LYY165 pKa = 10.76ADD167 pKa = 3.52WKK169 pKa = 9.79YY170 pKa = 10.4RR171 pKa = 11.84PEE173 pKa = 4.03VADD176 pKa = 3.67YY177 pKa = 8.97EE178 pKa = 4.6SPPGIFEE185 pKa = 4.11PVYY188 pKa = 11.09DD189 pKa = 3.77LTNWSEE195 pKa = 4.07ANLGRR200 pKa = 11.84VEE202 pKa = 3.92GRR204 pKa = 11.84PRR206 pKa = 11.84GLVLFGATRR215 pKa = 11.84LGKK218 pKa = 7.65TVWARR223 pKa = 11.84SLGKK227 pKa = 9.74HH228 pKa = 5.68NYY230 pKa = 8.93FAGLFNLDD238 pKa = 4.56DD239 pKa = 4.25FSEE242 pKa = 4.36SAEE245 pKa = 3.92YY246 pKa = 10.42AIFDD250 pKa = 4.19DD251 pKa = 4.0MSGGFSFFPSYY262 pKa = 9.2KK263 pKa = 9.6QWMGGQFQFTVTDD276 pKa = 3.65KK277 pKa = 10.77FKK279 pKa = 10.89HH280 pKa = 5.41KK281 pKa = 10.03RR282 pKa = 11.84TVRR285 pKa = 11.84WGKK288 pKa = 7.37PTIWLCNTDD297 pKa = 5.47PRR299 pKa = 11.84LDD301 pKa = 3.34WYY303 pKa = 10.98KK304 pKa = 10.58PGSSPDD310 pKa = 3.78FGWMEE315 pKa = 3.92EE316 pKa = 4.02NADD319 pKa = 3.77FVEE322 pKa = 4.27LTEE325 pKa = 4.52TIFRR329 pKa = 11.84ANRR332 pKa = 11.84EE333 pKa = 3.96
Molecular weight: 38.32 kDa
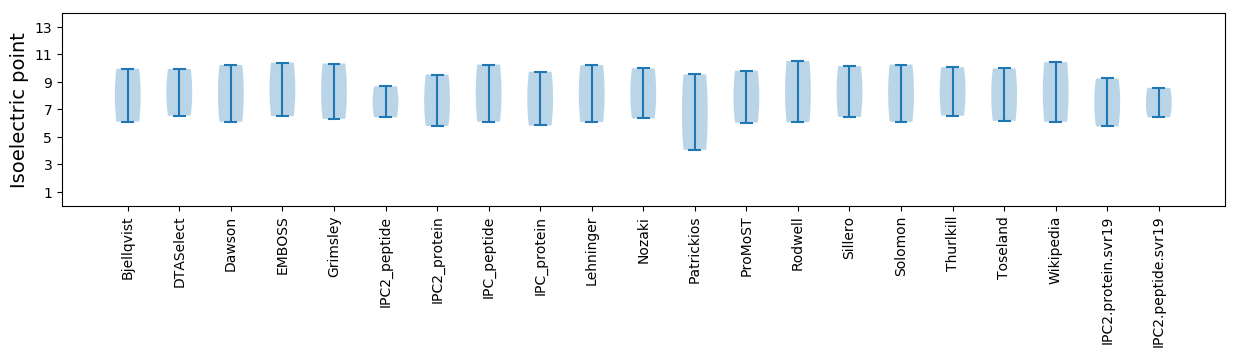
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BP67|A0A346BP67_9VIRU Putative capsid protein OS=Tubeweb spider associated circular virus 1 OX=2293308 PE=4 SV=1
MM1 pKa = 7.71AYY3 pKa = 10.24SRR5 pKa = 11.84ARR7 pKa = 11.84TYY9 pKa = 10.45RR10 pKa = 11.84RR11 pKa = 11.84PYY13 pKa = 10.05RR14 pKa = 11.84KK15 pKa = 9.38RR16 pKa = 11.84SGPKK20 pKa = 9.26SRR22 pKa = 11.84RR23 pKa = 11.84STGKK27 pKa = 9.47KK28 pKa = 6.78RR29 pKa = 11.84TYY31 pKa = 10.71RR32 pKa = 11.84KK33 pKa = 7.58TKK35 pKa = 8.11KK36 pKa = 8.64TSRR39 pKa = 11.84PMSNKK44 pKa = 9.89KK45 pKa = 9.88ILDD48 pKa = 3.29LTSRR52 pKa = 11.84KK53 pKa = 9.67KK54 pKa = 10.37RR55 pKa = 11.84DD56 pKa = 3.23TMAPYY61 pKa = 10.24YY62 pKa = 8.72NTYY65 pKa = 10.43GSSPLTGLGGYY76 pKa = 6.44TANGNGGGAQAYY88 pKa = 7.68TGLLWLATARR98 pKa = 11.84DD99 pKa = 3.7NTLASGGSVSNINDD113 pKa = 3.68LAARR117 pKa = 11.84TATACYY123 pKa = 9.63MRR125 pKa = 11.84GLKK128 pKa = 10.38EE129 pKa = 3.84NVEE132 pKa = 4.36ISTSSGAPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTFRR151 pKa = 11.84DD152 pKa = 4.06PSVVLSGTGIPPYY165 pKa = 10.77LEE167 pKa = 4.04TSNGYY172 pKa = 10.28ARR174 pKa = 11.84LLANFTNSDD183 pKa = 3.62SAGVATHH190 pKa = 6.53GAITRR195 pKa = 11.84LIFKK199 pKa = 8.95GQQNVDD205 pKa = 2.85WNDD208 pKa = 2.52IRR210 pKa = 11.84TAPIDD215 pKa = 3.58NARR218 pKa = 11.84VDD220 pKa = 4.01LKK222 pKa = 10.91YY223 pKa = 10.74DD224 pKa = 2.81KK225 pKa = 11.27TMIVSSGNANGRR237 pKa = 11.84VKK239 pKa = 10.41LFKK242 pKa = 9.42MWHH245 pKa = 6.16PMNKK249 pKa = 8.39TLVYY253 pKa = 10.81DD254 pKa = 4.05DD255 pKa = 5.29DD256 pKa = 4.93EE257 pKa = 6.01NGEE260 pKa = 4.4AEE262 pKa = 4.1NSTVFSVGDD271 pKa = 3.41KK272 pKa = 10.75RR273 pKa = 11.84GMGDD277 pKa = 3.45YY278 pKa = 10.86YY279 pKa = 11.49VIDD282 pKa = 4.2FLQAGMGATSSDD294 pKa = 3.51SLNFNPTATLYY305 pKa = 8.61WHH307 pKa = 6.59EE308 pKa = 4.22RR309 pKa = 3.47
MM1 pKa = 7.71AYY3 pKa = 10.24SRR5 pKa = 11.84ARR7 pKa = 11.84TYY9 pKa = 10.45RR10 pKa = 11.84RR11 pKa = 11.84PYY13 pKa = 10.05RR14 pKa = 11.84KK15 pKa = 9.38RR16 pKa = 11.84SGPKK20 pKa = 9.26SRR22 pKa = 11.84RR23 pKa = 11.84STGKK27 pKa = 9.47KK28 pKa = 6.78RR29 pKa = 11.84TYY31 pKa = 10.71RR32 pKa = 11.84KK33 pKa = 7.58TKK35 pKa = 8.11KK36 pKa = 8.64TSRR39 pKa = 11.84PMSNKK44 pKa = 9.89KK45 pKa = 9.88ILDD48 pKa = 3.29LTSRR52 pKa = 11.84KK53 pKa = 9.67KK54 pKa = 10.37RR55 pKa = 11.84DD56 pKa = 3.23TMAPYY61 pKa = 10.24YY62 pKa = 8.72NTYY65 pKa = 10.43GSSPLTGLGGYY76 pKa = 6.44TANGNGGGAQAYY88 pKa = 7.68TGLLWLATARR98 pKa = 11.84DD99 pKa = 3.7NTLASGGSVSNINDD113 pKa = 3.68LAARR117 pKa = 11.84TATACYY123 pKa = 9.63MRR125 pKa = 11.84GLKK128 pKa = 10.38EE129 pKa = 3.84NVEE132 pKa = 4.36ISTSSGAPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTFRR151 pKa = 11.84DD152 pKa = 4.06PSVVLSGTGIPPYY165 pKa = 10.77LEE167 pKa = 4.04TSNGYY172 pKa = 10.28ARR174 pKa = 11.84LLANFTNSDD183 pKa = 3.62SAGVATHH190 pKa = 6.53GAITRR195 pKa = 11.84LIFKK199 pKa = 8.95GQQNVDD205 pKa = 2.85WNDD208 pKa = 2.52IRR210 pKa = 11.84TAPIDD215 pKa = 3.58NARR218 pKa = 11.84VDD220 pKa = 4.01LKK222 pKa = 10.91YY223 pKa = 10.74DD224 pKa = 2.81KK225 pKa = 11.27TMIVSSGNANGRR237 pKa = 11.84VKK239 pKa = 10.41LFKK242 pKa = 9.42MWHH245 pKa = 6.16PMNKK249 pKa = 8.39TLVYY253 pKa = 10.81DD254 pKa = 4.05DD255 pKa = 5.29DD256 pKa = 4.93EE257 pKa = 6.01NGEE260 pKa = 4.4AEE262 pKa = 4.1NSTVFSVGDD271 pKa = 3.41KK272 pKa = 10.75RR273 pKa = 11.84GMGDD277 pKa = 3.45YY278 pKa = 10.86YY279 pKa = 11.49VIDD282 pKa = 4.2FLQAGMGATSSDD294 pKa = 3.51SLNFNPTATLYY305 pKa = 8.61WHH307 pKa = 6.59EE308 pKa = 4.22RR309 pKa = 3.47
Molecular weight: 34.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
642 |
309 |
333 |
321.0 |
36.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.854 ± 1.027 | 1.246 ± 0.394 |
6.386 ± 0.369 | 4.361 ± 1.379 |
4.829 ± 1.474 | 9.813 ± 0.282 |
1.713 ± 0.489 | 3.583 ± 0.015 |
5.763 ± 0.254 | 6.854 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.336 ± 0.379 | 5.452 ± 0.885 |
4.517 ± 0.417 | 2.181 ± 0.37 |
7.477 ± 0.617 | 6.854 ± 1.24 |
7.009 ± 1.563 | 4.829 ± 0.409 |
2.804 ± 0.567 | 5.14 ± 0.238 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
