
Metschnikowia sp. JCM 33374
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Metschnikowiaceae; Metschnikowia; unclassified Metschnikowia
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
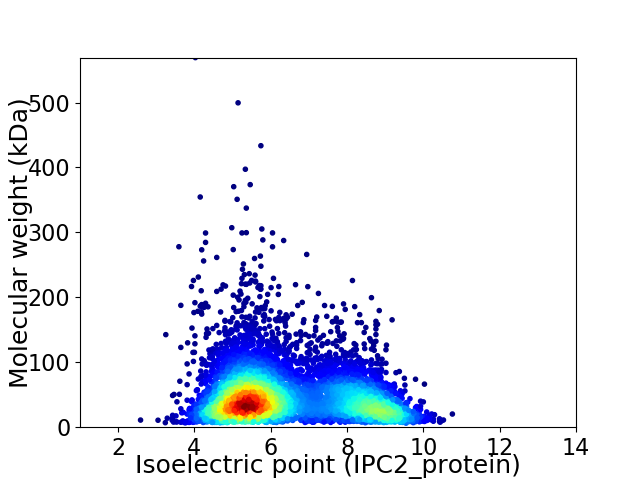
Virtual 2D-PAGE plot for 6652 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A512U876|A0A512U876_9ASCO Uncharacterized protein OS=Metschnikowia sp. JCM 33374 OX=2562755 GN=JCM33374_g943 PE=4 SV=1
MM1 pKa = 7.54GSAPSDD7 pKa = 3.74SIFNDD12 pKa = 3.38VLDD15 pKa = 4.59GNPPGEE21 pKa = 4.41SIKK24 pKa = 10.85PPFPTATSMGEE35 pKa = 4.0RR36 pKa = 11.84IVGLNNRR43 pKa = 11.84EE44 pKa = 4.06EE45 pKa = 4.18KK46 pKa = 10.55CIQGEE51 pKa = 4.28TRR53 pKa = 11.84ATNEE57 pKa = 3.64VGKK60 pKa = 10.42ADD62 pKa = 4.26HH63 pKa = 6.76LVEE66 pKa = 4.58HH67 pKa = 6.79CPIDD71 pKa = 4.0EE72 pKa = 4.81DD73 pKa = 5.29SSDD76 pKa = 3.77EE77 pKa = 4.07EE78 pKa = 4.56NINLIPRR85 pKa = 11.84CKK87 pKa = 10.33FIITSQDD94 pKa = 3.44DD95 pKa = 3.78NCTPEE100 pKa = 4.35AVGNGTPNDD109 pKa = 3.78YY110 pKa = 9.82TSSSEE115 pKa = 3.96TPEE118 pKa = 4.95IIGQVYY124 pKa = 9.78LVQSDD129 pKa = 3.72NDD131 pKa = 4.03CYY133 pKa = 10.93RR134 pKa = 11.84SDD136 pKa = 3.51VLFEE140 pKa = 4.68GFSSYY145 pKa = 7.24QTVNRR150 pKa = 11.84NQPRR154 pKa = 11.84RR155 pKa = 11.84TTKK158 pKa = 10.53NSINAFRR165 pKa = 11.84RR166 pKa = 11.84SQNGHH171 pKa = 5.36FSYY174 pKa = 11.07YY175 pKa = 10.05EE176 pKa = 4.09SPHH179 pKa = 6.38EE180 pKa = 4.26EE181 pKa = 4.27SQSSDD186 pKa = 2.96SVYY189 pKa = 11.26DD190 pKa = 3.49EE191 pKa = 6.01DD192 pKa = 5.21EE193 pKa = 4.6FSTEE197 pKa = 3.74QAIDD201 pKa = 3.26EE202 pKa = 4.49GSPLKK207 pKa = 10.6KK208 pKa = 9.77DD209 pKa = 3.25TSYY212 pKa = 11.58SCDD215 pKa = 3.98PIYY218 pKa = 10.76LLSDD222 pKa = 3.35EE223 pKa = 4.73TTLSDD228 pKa = 3.93DD229 pKa = 3.64YY230 pKa = 11.86SYY232 pKa = 11.93GDD234 pKa = 3.51SNTYY238 pKa = 10.6GNSSRR243 pKa = 11.84PYY245 pKa = 10.05IPDD248 pKa = 3.44STSDD252 pKa = 3.64SQDD255 pKa = 3.12STDD258 pKa = 3.97SQDD261 pKa = 3.4STDD264 pKa = 4.01SQDD267 pKa = 3.4STDD270 pKa = 4.01SQDD273 pKa = 3.4STDD276 pKa = 4.01SQDD279 pKa = 3.45STDD282 pKa = 3.66SQVPTIEE289 pKa = 4.71DD290 pKa = 3.54TTSDD294 pKa = 4.28DD295 pKa = 3.93NEE297 pKa = 4.63SMDD300 pKa = 3.54TYY302 pKa = 11.55VYY304 pKa = 10.37ISSKK308 pKa = 8.58EE309 pKa = 3.88HH310 pKa = 5.7SSSYY314 pKa = 9.93VAEE317 pKa = 4.66DD318 pKa = 5.27DD319 pKa = 4.27IVLDD323 pKa = 4.13PEE325 pKa = 4.47DD326 pKa = 4.76EE327 pKa = 4.32EE328 pKa = 4.84VLSQEE333 pKa = 4.85KK334 pKa = 10.53YY335 pKa = 6.79PTPPEE340 pKa = 4.38PPNPPDD346 pKa = 3.82PSNPPEE352 pKa = 4.32PPKK355 pKa = 10.6PPVPNRR361 pKa = 11.84YY362 pKa = 8.81KK363 pKa = 10.79ASPNSFPQSQEE374 pKa = 3.89TPP376 pKa = 3.3
MM1 pKa = 7.54GSAPSDD7 pKa = 3.74SIFNDD12 pKa = 3.38VLDD15 pKa = 4.59GNPPGEE21 pKa = 4.41SIKK24 pKa = 10.85PPFPTATSMGEE35 pKa = 4.0RR36 pKa = 11.84IVGLNNRR43 pKa = 11.84EE44 pKa = 4.06EE45 pKa = 4.18KK46 pKa = 10.55CIQGEE51 pKa = 4.28TRR53 pKa = 11.84ATNEE57 pKa = 3.64VGKK60 pKa = 10.42ADD62 pKa = 4.26HH63 pKa = 6.76LVEE66 pKa = 4.58HH67 pKa = 6.79CPIDD71 pKa = 4.0EE72 pKa = 4.81DD73 pKa = 5.29SSDD76 pKa = 3.77EE77 pKa = 4.07EE78 pKa = 4.56NINLIPRR85 pKa = 11.84CKK87 pKa = 10.33FIITSQDD94 pKa = 3.44DD95 pKa = 3.78NCTPEE100 pKa = 4.35AVGNGTPNDD109 pKa = 3.78YY110 pKa = 9.82TSSSEE115 pKa = 3.96TPEE118 pKa = 4.95IIGQVYY124 pKa = 9.78LVQSDD129 pKa = 3.72NDD131 pKa = 4.03CYY133 pKa = 10.93RR134 pKa = 11.84SDD136 pKa = 3.51VLFEE140 pKa = 4.68GFSSYY145 pKa = 7.24QTVNRR150 pKa = 11.84NQPRR154 pKa = 11.84RR155 pKa = 11.84TTKK158 pKa = 10.53NSINAFRR165 pKa = 11.84RR166 pKa = 11.84SQNGHH171 pKa = 5.36FSYY174 pKa = 11.07YY175 pKa = 10.05EE176 pKa = 4.09SPHH179 pKa = 6.38EE180 pKa = 4.26EE181 pKa = 4.27SQSSDD186 pKa = 2.96SVYY189 pKa = 11.26DD190 pKa = 3.49EE191 pKa = 6.01DD192 pKa = 5.21EE193 pKa = 4.6FSTEE197 pKa = 3.74QAIDD201 pKa = 3.26EE202 pKa = 4.49GSPLKK207 pKa = 10.6KK208 pKa = 9.77DD209 pKa = 3.25TSYY212 pKa = 11.58SCDD215 pKa = 3.98PIYY218 pKa = 10.76LLSDD222 pKa = 3.35EE223 pKa = 4.73TTLSDD228 pKa = 3.93DD229 pKa = 3.64YY230 pKa = 11.86SYY232 pKa = 11.93GDD234 pKa = 3.51SNTYY238 pKa = 10.6GNSSRR243 pKa = 11.84PYY245 pKa = 10.05IPDD248 pKa = 3.44STSDD252 pKa = 3.64SQDD255 pKa = 3.12STDD258 pKa = 3.97SQDD261 pKa = 3.4STDD264 pKa = 4.01SQDD267 pKa = 3.4STDD270 pKa = 4.01SQDD273 pKa = 3.4STDD276 pKa = 4.01SQDD279 pKa = 3.45STDD282 pKa = 3.66SQVPTIEE289 pKa = 4.71DD290 pKa = 3.54TTSDD294 pKa = 4.28DD295 pKa = 3.93NEE297 pKa = 4.63SMDD300 pKa = 3.54TYY302 pKa = 11.55VYY304 pKa = 10.37ISSKK308 pKa = 8.58EE309 pKa = 3.88HH310 pKa = 5.7SSSYY314 pKa = 9.93VAEE317 pKa = 4.66DD318 pKa = 5.27DD319 pKa = 4.27IVLDD323 pKa = 4.13PEE325 pKa = 4.47DD326 pKa = 4.76EE327 pKa = 4.32EE328 pKa = 4.84VLSQEE333 pKa = 4.85KK334 pKa = 10.53YY335 pKa = 6.79PTPPEE340 pKa = 4.38PPNPPDD346 pKa = 3.82PSNPPEE352 pKa = 4.32PPKK355 pKa = 10.6PPVPNRR361 pKa = 11.84YY362 pKa = 8.81KK363 pKa = 10.79ASPNSFPQSQEE374 pKa = 3.89TPP376 pKa = 3.3
Molecular weight: 41.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A512UGH6|A0A512UGH6_9ASCO Uncharacterized protein OS=Metschnikowia sp. JCM 33374 OX=2562755 GN=JCM33374_g3829 PE=4 SV=1
MM1 pKa = 7.83ANRR4 pKa = 11.84TVGSCVGSSGKK15 pKa = 9.67SSRR18 pKa = 11.84SLTICVRR25 pKa = 11.84AHH27 pKa = 6.38NGHH30 pKa = 5.91SLRR33 pKa = 11.84GGVRR37 pKa = 11.84TGQSRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84EE45 pKa = 4.27WNNHH49 pKa = 3.48NRR51 pKa = 11.84SLGSISAAYY60 pKa = 9.61SLSGGVRR67 pKa = 11.84SCPSRR72 pKa = 11.84GGEE75 pKa = 4.01WNNHH79 pKa = 3.81NRR81 pKa = 11.84SLGSISAGQQEE92 pKa = 5.37CKK94 pKa = 10.2NQSKK98 pKa = 10.27VEE100 pKa = 3.99VEE102 pKa = 4.12EE103 pKa = 3.99TRR105 pKa = 11.84NNHH108 pKa = 5.47TGVLAPSGGYY118 pKa = 7.05SQRR121 pKa = 11.84VVVRR125 pKa = 11.84TRR127 pKa = 11.84EE128 pKa = 4.01VEE130 pKa = 4.16EE131 pKa = 4.0VRR133 pKa = 11.84NNHH136 pKa = 4.66NWRR139 pKa = 11.84LGMEE143 pKa = 4.35QSTKK147 pKa = 11.1DD148 pKa = 3.48PVQVEE153 pKa = 4.54VEE155 pKa = 4.13NGTTTTGVLAPSVSLSGGKK174 pKa = 9.41ISSKK178 pKa = 11.02
MM1 pKa = 7.83ANRR4 pKa = 11.84TVGSCVGSSGKK15 pKa = 9.67SSRR18 pKa = 11.84SLTICVRR25 pKa = 11.84AHH27 pKa = 6.38NGHH30 pKa = 5.91SLRR33 pKa = 11.84GGVRR37 pKa = 11.84TGQSRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84EE45 pKa = 4.27WNNHH49 pKa = 3.48NRR51 pKa = 11.84SLGSISAAYY60 pKa = 9.61SLSGGVRR67 pKa = 11.84SCPSRR72 pKa = 11.84GGEE75 pKa = 4.01WNNHH79 pKa = 3.81NRR81 pKa = 11.84SLGSISAGQQEE92 pKa = 5.37CKK94 pKa = 10.2NQSKK98 pKa = 10.27VEE100 pKa = 3.99VEE102 pKa = 4.12EE103 pKa = 3.99TRR105 pKa = 11.84NNHH108 pKa = 5.47TGVLAPSGGYY118 pKa = 7.05SQRR121 pKa = 11.84VVVRR125 pKa = 11.84TRR127 pKa = 11.84EE128 pKa = 4.01VEE130 pKa = 4.16EE131 pKa = 4.0VRR133 pKa = 11.84NNHH136 pKa = 4.66NWRR139 pKa = 11.84LGMEE143 pKa = 4.35QSTKK147 pKa = 11.1DD148 pKa = 3.48PVQVEE153 pKa = 4.54VEE155 pKa = 4.13NGTTTTGVLAPSVSLSGGKK174 pKa = 9.41ISSKK178 pKa = 11.02
Molecular weight: 19.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3060062 |
66 |
5577 |
460.0 |
51.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.687 ± 0.033 | 1.186 ± 0.012 |
5.712 ± 0.026 | 6.418 ± 0.035 |
4.389 ± 0.023 | 5.646 ± 0.034 |
2.364 ± 0.014 | 5.494 ± 0.025 |
6.417 ± 0.036 | 8.869 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.015 ± 0.013 | 4.789 ± 0.022 |
5.001 ± 0.031 | 3.881 ± 0.025 |
4.481 ± 0.024 | 9.716 ± 0.055 |
6.324 ± 0.094 | 6.386 ± 0.036 |
1.078 ± 0.012 | 3.15 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
