
Luminiphilus syltensis NOR5-1B
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Luminiphilus; Luminiphilus syltensis
Average proteome isoelectric point is 5.75
Get precalculated fractions of proteins
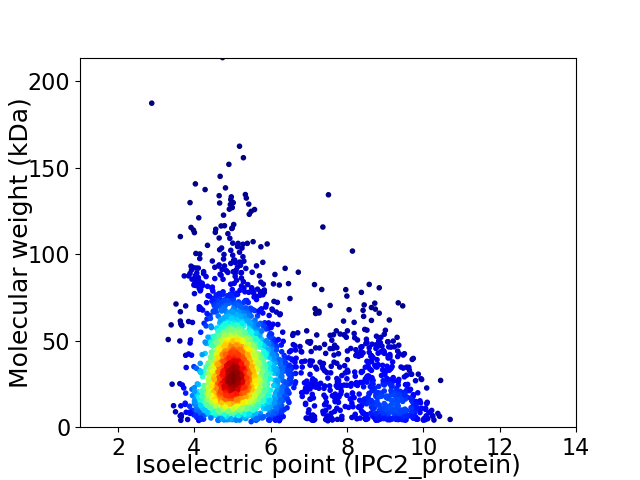
Virtual 2D-PAGE plot for 2923 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B8KTG7|B8KTG7_9GAMM FOG: CBS domain protein OS=Luminiphilus syltensis NOR5-1B OX=565045 GN=NOR51B_1511 PE=4 SV=1
MM1 pKa = 7.2GQFDD5 pKa = 4.09SEE7 pKa = 4.58ANAMRR12 pKa = 11.84YY13 pKa = 7.79VAEE16 pKa = 4.47GAFGGPLSEE25 pKa = 4.21TVAARR30 pKa = 11.84AAFRR34 pKa = 11.84YY35 pKa = 9.51SKK37 pKa = 10.52QDD39 pKa = 3.25PYY41 pKa = 11.71LKK43 pKa = 10.43NLYY46 pKa = 10.78SNDD49 pKa = 3.55DD50 pKa = 3.57QLPFLGDD57 pKa = 3.76PGPGAGADD65 pKa = 4.03LGDD68 pKa = 5.55DD69 pKa = 3.61DD70 pKa = 5.49TIAARR75 pKa = 11.84LRR77 pKa = 11.84FAFTPSDD84 pKa = 4.2DD85 pKa = 4.04LRR87 pKa = 11.84MDD89 pKa = 4.13LSFNYY94 pKa = 9.94SDD96 pKa = 4.5SEE98 pKa = 4.76VNTGPYY104 pKa = 9.3QSKK107 pKa = 8.57STLGIATEE115 pKa = 4.26VNGNLEE121 pKa = 4.9LINVVNTPADD131 pKa = 3.88EE132 pKa = 4.37TRR134 pKa = 11.84LTAVVDD140 pKa = 3.66ADD142 pKa = 4.4GNDD145 pKa = 3.24TGLDD149 pKa = 3.29GGADD153 pKa = 3.46IEE155 pKa = 5.3DD156 pKa = 4.14GDD158 pKa = 4.33SFLPGGGGGLPGRR171 pKa = 11.84LAPGADD177 pKa = 3.17FFGYY181 pKa = 10.46LDD183 pKa = 4.33PDD185 pKa = 4.03GDD187 pKa = 4.54DD188 pKa = 3.71FTFSGDD194 pKa = 3.28FAFRR198 pKa = 11.84DD199 pKa = 3.47QGSVEE204 pKa = 3.79NSGVNFRR211 pKa = 11.84LIYY214 pKa = 10.35DD215 pKa = 4.37LSDD218 pKa = 3.11TTTFTSITDD227 pKa = 3.47YY228 pKa = 11.4KK229 pKa = 11.0DD230 pKa = 3.37YY231 pKa = 11.31EE232 pKa = 4.16KK233 pKa = 11.32LLFIDD238 pKa = 3.91VDD240 pKa = 3.87AAPVNQLANYY250 pKa = 9.4AGVDD254 pKa = 3.34ASSFTQEE261 pKa = 3.66FRR263 pKa = 11.84LSGEE267 pKa = 4.31TEE269 pKa = 3.63RR270 pKa = 11.84SRR272 pKa = 11.84WVAGLYY278 pKa = 9.63YY279 pKa = 10.79LNIDD283 pKa = 3.87SEE285 pKa = 4.69SDD287 pKa = 3.23NGLKK291 pKa = 10.61GPQNSFADD299 pKa = 3.79VFGGSPIDD307 pKa = 3.65VGTVANLEE315 pKa = 4.11TDD317 pKa = 3.32SYY319 pKa = 11.58SIFGQYY325 pKa = 10.42EE326 pKa = 4.0YY327 pKa = 11.34DD328 pKa = 3.9FSDD331 pKa = 3.66KK332 pKa = 10.43LTVIAGLRR340 pKa = 11.84AMQEE344 pKa = 3.94EE345 pKa = 4.81KK346 pKa = 11.11DD347 pKa = 3.62FGLVLGVAPSDD358 pKa = 3.47GSRR361 pKa = 11.84VVNSSPCFSGQLDD374 pKa = 3.55ACFPGTGVSYY384 pKa = 11.23EE385 pKa = 4.53DD386 pKa = 4.61DD387 pKa = 3.65FSEE390 pKa = 4.59NFWAGKK396 pKa = 9.29LQLTYY401 pKa = 10.83DD402 pKa = 3.6VSDD405 pKa = 4.63DD406 pKa = 3.48LMLYY410 pKa = 10.42AGVNRR415 pKa = 11.84GVKK418 pKa = 9.82AGSYY422 pKa = 8.46NAPLLGAYY430 pKa = 9.47FGSGGDD436 pKa = 3.12AGLPYY441 pKa = 10.62DD442 pKa = 4.24EE443 pKa = 5.6EE444 pKa = 4.56VLTSYY449 pKa = 10.91EE450 pKa = 4.28GGFKK454 pKa = 9.91ATLGGGTTRR463 pKa = 11.84INATAFYY470 pKa = 10.29YY471 pKa = 10.14DD472 pKa = 3.65YY473 pKa = 11.36KK474 pKa = 10.99DD475 pKa = 3.49YY476 pKa = 11.31QAFLFVGVGGVVTNQDD492 pKa = 2.77ADD494 pKa = 3.59NYY496 pKa = 10.13GVEE499 pKa = 5.22LEE501 pKa = 4.3IQTTPVDD508 pKa = 3.92GLDD511 pKa = 4.27LIISGAYY518 pKa = 9.65FDD520 pKa = 4.68ATVKK524 pKa = 10.68DD525 pKa = 3.66VSLRR529 pKa = 11.84NGSPLPPSDD538 pKa = 4.27VDD540 pKa = 3.28PTYY543 pKa = 11.16APEE546 pKa = 4.28LQATALARR554 pKa = 11.84YY555 pKa = 9.85SFDD558 pKa = 3.51AFGGLMAIQGDD569 pKa = 3.38ISYY572 pKa = 11.0SDD574 pKa = 3.5EE575 pKa = 4.09YY576 pKa = 11.39YY577 pKa = 10.95YY578 pKa = 11.26NLRR581 pKa = 11.84NFDD584 pKa = 3.51ADD586 pKa = 3.53KK587 pKa = 10.64FDD589 pKa = 4.43SYY591 pKa = 11.67VVANAQLSYY600 pKa = 11.38AVDD603 pKa = 3.28DD604 pKa = 4.14WLATLAVRR612 pKa = 11.84NLTDD616 pKa = 3.41EE617 pKa = 4.21RR618 pKa = 11.84VGIQGFDD625 pKa = 3.25LATLCGCNEE634 pKa = 4.06VAYY637 pKa = 9.59KK638 pKa = 10.17APRR641 pKa = 11.84YY642 pKa = 9.67FSFGLRR648 pKa = 11.84KK649 pKa = 10.23DD650 pKa = 3.58FF651 pKa = 4.77
MM1 pKa = 7.2GQFDD5 pKa = 4.09SEE7 pKa = 4.58ANAMRR12 pKa = 11.84YY13 pKa = 7.79VAEE16 pKa = 4.47GAFGGPLSEE25 pKa = 4.21TVAARR30 pKa = 11.84AAFRR34 pKa = 11.84YY35 pKa = 9.51SKK37 pKa = 10.52QDD39 pKa = 3.25PYY41 pKa = 11.71LKK43 pKa = 10.43NLYY46 pKa = 10.78SNDD49 pKa = 3.55DD50 pKa = 3.57QLPFLGDD57 pKa = 3.76PGPGAGADD65 pKa = 4.03LGDD68 pKa = 5.55DD69 pKa = 3.61DD70 pKa = 5.49TIAARR75 pKa = 11.84LRR77 pKa = 11.84FAFTPSDD84 pKa = 4.2DD85 pKa = 4.04LRR87 pKa = 11.84MDD89 pKa = 4.13LSFNYY94 pKa = 9.94SDD96 pKa = 4.5SEE98 pKa = 4.76VNTGPYY104 pKa = 9.3QSKK107 pKa = 8.57STLGIATEE115 pKa = 4.26VNGNLEE121 pKa = 4.9LINVVNTPADD131 pKa = 3.88EE132 pKa = 4.37TRR134 pKa = 11.84LTAVVDD140 pKa = 3.66ADD142 pKa = 4.4GNDD145 pKa = 3.24TGLDD149 pKa = 3.29GGADD153 pKa = 3.46IEE155 pKa = 5.3DD156 pKa = 4.14GDD158 pKa = 4.33SFLPGGGGGLPGRR171 pKa = 11.84LAPGADD177 pKa = 3.17FFGYY181 pKa = 10.46LDD183 pKa = 4.33PDD185 pKa = 4.03GDD187 pKa = 4.54DD188 pKa = 3.71FTFSGDD194 pKa = 3.28FAFRR198 pKa = 11.84DD199 pKa = 3.47QGSVEE204 pKa = 3.79NSGVNFRR211 pKa = 11.84LIYY214 pKa = 10.35DD215 pKa = 4.37LSDD218 pKa = 3.11TTTFTSITDD227 pKa = 3.47YY228 pKa = 11.4KK229 pKa = 11.0DD230 pKa = 3.37YY231 pKa = 11.31EE232 pKa = 4.16KK233 pKa = 11.32LLFIDD238 pKa = 3.91VDD240 pKa = 3.87AAPVNQLANYY250 pKa = 9.4AGVDD254 pKa = 3.34ASSFTQEE261 pKa = 3.66FRR263 pKa = 11.84LSGEE267 pKa = 4.31TEE269 pKa = 3.63RR270 pKa = 11.84SRR272 pKa = 11.84WVAGLYY278 pKa = 9.63YY279 pKa = 10.79LNIDD283 pKa = 3.87SEE285 pKa = 4.69SDD287 pKa = 3.23NGLKK291 pKa = 10.61GPQNSFADD299 pKa = 3.79VFGGSPIDD307 pKa = 3.65VGTVANLEE315 pKa = 4.11TDD317 pKa = 3.32SYY319 pKa = 11.58SIFGQYY325 pKa = 10.42EE326 pKa = 4.0YY327 pKa = 11.34DD328 pKa = 3.9FSDD331 pKa = 3.66KK332 pKa = 10.43LTVIAGLRR340 pKa = 11.84AMQEE344 pKa = 3.94EE345 pKa = 4.81KK346 pKa = 11.11DD347 pKa = 3.62FGLVLGVAPSDD358 pKa = 3.47GSRR361 pKa = 11.84VVNSSPCFSGQLDD374 pKa = 3.55ACFPGTGVSYY384 pKa = 11.23EE385 pKa = 4.53DD386 pKa = 4.61DD387 pKa = 3.65FSEE390 pKa = 4.59NFWAGKK396 pKa = 9.29LQLTYY401 pKa = 10.83DD402 pKa = 3.6VSDD405 pKa = 4.63DD406 pKa = 3.48LMLYY410 pKa = 10.42AGVNRR415 pKa = 11.84GVKK418 pKa = 9.82AGSYY422 pKa = 8.46NAPLLGAYY430 pKa = 9.47FGSGGDD436 pKa = 3.12AGLPYY441 pKa = 10.62DD442 pKa = 4.24EE443 pKa = 5.6EE444 pKa = 4.56VLTSYY449 pKa = 10.91EE450 pKa = 4.28GGFKK454 pKa = 9.91ATLGGGTTRR463 pKa = 11.84INATAFYY470 pKa = 10.29YY471 pKa = 10.14DD472 pKa = 3.65YY473 pKa = 11.36KK474 pKa = 10.99DD475 pKa = 3.49YY476 pKa = 11.31QAFLFVGVGGVVTNQDD492 pKa = 2.77ADD494 pKa = 3.59NYY496 pKa = 10.13GVEE499 pKa = 5.22LEE501 pKa = 4.3IQTTPVDD508 pKa = 3.92GLDD511 pKa = 4.27LIISGAYY518 pKa = 9.65FDD520 pKa = 4.68ATVKK524 pKa = 10.68DD525 pKa = 3.66VSLRR529 pKa = 11.84NGSPLPPSDD538 pKa = 4.27VDD540 pKa = 3.28PTYY543 pKa = 11.16APEE546 pKa = 4.28LQATALARR554 pKa = 11.84YY555 pKa = 9.85SFDD558 pKa = 3.51AFGGLMAIQGDD569 pKa = 3.38ISYY572 pKa = 11.0SDD574 pKa = 3.5EE575 pKa = 4.09YY576 pKa = 11.39YY577 pKa = 10.95YY578 pKa = 11.26NLRR581 pKa = 11.84NFDD584 pKa = 3.51ADD586 pKa = 3.53KK587 pKa = 10.64FDD589 pKa = 4.43SYY591 pKa = 11.67VVANAQLSYY600 pKa = 11.38AVDD603 pKa = 3.28DD604 pKa = 4.14WLATLAVRR612 pKa = 11.84NLTDD616 pKa = 3.41EE617 pKa = 4.21RR618 pKa = 11.84VGIQGFDD625 pKa = 3.25LATLCGCNEE634 pKa = 4.06VAYY637 pKa = 9.59KK638 pKa = 10.17APRR641 pKa = 11.84YY642 pKa = 9.67FSFGLRR648 pKa = 11.84KK649 pKa = 10.23DD650 pKa = 3.58FF651 pKa = 4.77
Molecular weight: 70.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B8KXL6|B8KXL6_9GAMM Long-chain-fatty-acid--CoA ligase OS=Luminiphilus syltensis NOR5-1B OX=565045 GN=fadD PE=4 SV=1
MM1 pKa = 7.56NFRR4 pKa = 11.84KK5 pKa = 10.21ALDD8 pKa = 3.44AKK10 pKa = 9.75IQRR13 pKa = 11.84EE14 pKa = 3.39IAYY17 pKa = 9.77RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84FRR22 pKa = 11.84RR23 pKa = 11.84WLVYY27 pKa = 9.99FSLSCLCWTTGMAIPFFTLNTSGSFRR53 pKa = 11.84GALLAIYY60 pKa = 9.18FAGVALLVVWSDD72 pKa = 3.08RR73 pKa = 11.84KK74 pKa = 10.47SRR76 pKa = 11.84QSNDD80 pKa = 3.19DD81 pKa = 3.15
MM1 pKa = 7.56NFRR4 pKa = 11.84KK5 pKa = 10.21ALDD8 pKa = 3.44AKK10 pKa = 9.75IQRR13 pKa = 11.84EE14 pKa = 3.39IAYY17 pKa = 9.77RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84FRR22 pKa = 11.84RR23 pKa = 11.84WLVYY27 pKa = 9.99FSLSCLCWTTGMAIPFFTLNTSGSFRR53 pKa = 11.84GALLAIYY60 pKa = 9.18FAGVALLVVWSDD72 pKa = 3.08RR73 pKa = 11.84KK74 pKa = 10.47SRR76 pKa = 11.84QSNDD80 pKa = 3.19DD81 pKa = 3.15
Molecular weight: 9.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
948102 |
30 |
1947 |
324.4 |
35.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.65 ± 0.051 | 1.006 ± 0.015 |
6.284 ± 0.038 | 6.242 ± 0.043 |
3.779 ± 0.031 | 8.132 ± 0.041 |
2.085 ± 0.026 | 5.32 ± 0.028 |
3.058 ± 0.038 | 10.252 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.021 | 3.09 ± 0.028 |
4.819 ± 0.029 | 3.608 ± 0.024 |
6.336 ± 0.043 | 6.223 ± 0.036 |
5.4 ± 0.041 | 7.365 ± 0.038 |
1.354 ± 0.019 | 2.612 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
