
Nocardiopsis sp. CNR-923
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Nocardiopsis; unclassified Nocardiopsis
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
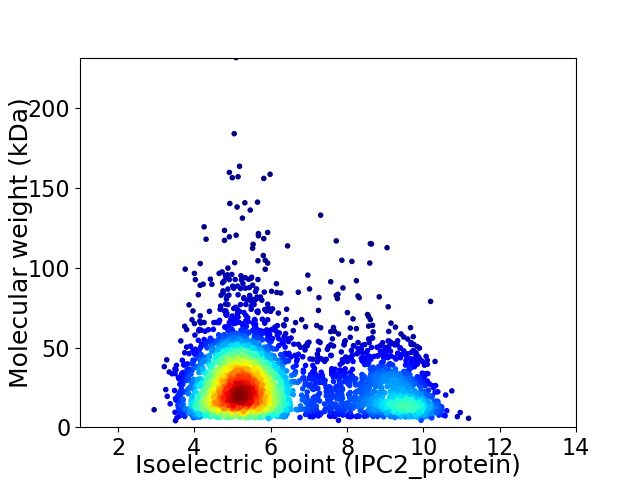
Virtual 2D-PAGE plot for 3756 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
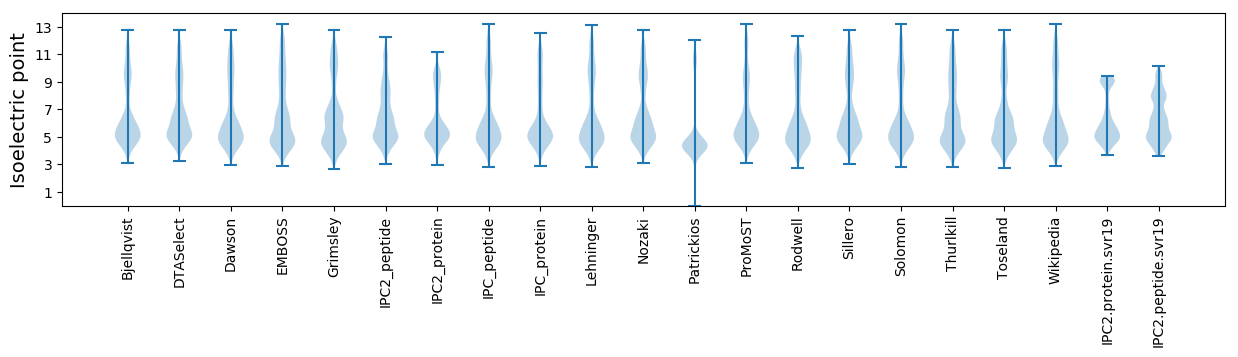
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q9TMI5|A0A1Q9TMI5_9ACTN Uncharacterized protein OS=Nocardiopsis sp. CNR-923 OX=1904965 GN=BJF83_20670 PE=4 SV=1
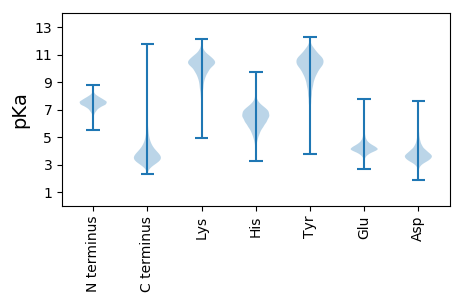
MM1 pKa = 7.07TRR3 pKa = 11.84HH4 pKa = 5.43PVARR8 pKa = 11.84SLGTTALSLTLVLAGAGLVSADD30 pKa = 3.67TTTGDD35 pKa = 3.31ISPEE39 pKa = 3.37QMAAMRR45 pKa = 11.84AAFGLSEE52 pKa = 4.61SGVDD56 pKa = 4.94DD57 pKa = 4.37LFEE60 pKa = 4.42AQEE63 pKa = 4.07EE64 pKa = 4.4ATEE67 pKa = 4.3TEE69 pKa = 3.96AEE71 pKa = 4.04LRR73 pKa = 11.84EE74 pKa = 4.16DD75 pKa = 3.71LGADD79 pKa = 3.62FGGAFFDD86 pKa = 5.14VDD88 pKa = 3.97TLDD91 pKa = 3.68LTVQVTDD98 pKa = 4.84PSASDD103 pKa = 3.39AVRR106 pKa = 11.84DD107 pKa = 3.89AGAVPEE113 pKa = 4.25VVDD116 pKa = 3.67NGEE119 pKa = 4.18AEE121 pKa = 4.05LAAGVEE127 pKa = 4.22ALDD130 pKa = 4.95AVDD133 pKa = 4.53EE134 pKa = 4.73VPATVHH140 pKa = 5.23GWYY143 pKa = 10.38ADD145 pKa = 3.71TVRR148 pKa = 11.84DD149 pKa = 3.63AVVIEE154 pKa = 4.14AAPGHH159 pKa = 5.52EE160 pKa = 4.08AEE162 pKa = 4.61ARR164 pKa = 11.84ALAEE168 pKa = 4.07RR169 pKa = 11.84AGVEE173 pKa = 3.51ADD175 pKa = 3.67TVVEE179 pKa = 4.25EE180 pKa = 4.68SVGTPTTYY188 pKa = 11.51ADD190 pKa = 3.09IVGGNPYY197 pKa = 10.33YY198 pKa = 10.38FQQDD202 pKa = 3.61GGWYY206 pKa = 7.85VCSVGFGVVGGYY218 pKa = 7.49VTAGHH223 pKa = 6.65CGDD226 pKa = 4.61EE227 pKa = 5.87GSDD230 pKa = 3.1TWLDD234 pKa = 3.57VPGTQQIGTVAGSSFPTSDD253 pKa = 3.66SAWVEE258 pKa = 4.11VTGSSTVPTAEE269 pKa = 3.92VNDD272 pKa = 4.11YY273 pKa = 11.25EE274 pKa = 5.19GGTVTVTGSTEE285 pKa = 4.02APVGASVCRR294 pKa = 11.84SGQTTGWHH302 pKa = 6.12CGVIEE307 pKa = 5.26AKK309 pKa = 10.48DD310 pKa = 3.32QTVRR314 pKa = 11.84YY315 pKa = 9.23AGNLVVYY322 pKa = 10.27GLTRR326 pKa = 11.84TSACAEE332 pKa = 4.31GGDD335 pKa = 5.2SGGSWLAGTEE345 pKa = 4.19AQGVTSGGSGNCTFGGTTFFQPLNPILDD373 pKa = 3.82EE374 pKa = 4.24WDD376 pKa = 3.38LTLLTGG382 pKa = 4.58
MM1 pKa = 7.07TRR3 pKa = 11.84HH4 pKa = 5.43PVARR8 pKa = 11.84SLGTTALSLTLVLAGAGLVSADD30 pKa = 3.67TTTGDD35 pKa = 3.31ISPEE39 pKa = 3.37QMAAMRR45 pKa = 11.84AAFGLSEE52 pKa = 4.61SGVDD56 pKa = 4.94DD57 pKa = 4.37LFEE60 pKa = 4.42AQEE63 pKa = 4.07EE64 pKa = 4.4ATEE67 pKa = 4.3TEE69 pKa = 3.96AEE71 pKa = 4.04LRR73 pKa = 11.84EE74 pKa = 4.16DD75 pKa = 3.71LGADD79 pKa = 3.62FGGAFFDD86 pKa = 5.14VDD88 pKa = 3.97TLDD91 pKa = 3.68LTVQVTDD98 pKa = 4.84PSASDD103 pKa = 3.39AVRR106 pKa = 11.84DD107 pKa = 3.89AGAVPEE113 pKa = 4.25VVDD116 pKa = 3.67NGEE119 pKa = 4.18AEE121 pKa = 4.05LAAGVEE127 pKa = 4.22ALDD130 pKa = 4.95AVDD133 pKa = 4.53EE134 pKa = 4.73VPATVHH140 pKa = 5.23GWYY143 pKa = 10.38ADD145 pKa = 3.71TVRR148 pKa = 11.84DD149 pKa = 3.63AVVIEE154 pKa = 4.14AAPGHH159 pKa = 5.52EE160 pKa = 4.08AEE162 pKa = 4.61ARR164 pKa = 11.84ALAEE168 pKa = 4.07RR169 pKa = 11.84AGVEE173 pKa = 3.51ADD175 pKa = 3.67TVVEE179 pKa = 4.25EE180 pKa = 4.68SVGTPTTYY188 pKa = 11.51ADD190 pKa = 3.09IVGGNPYY197 pKa = 10.33YY198 pKa = 10.38FQQDD202 pKa = 3.61GGWYY206 pKa = 7.85VCSVGFGVVGGYY218 pKa = 7.49VTAGHH223 pKa = 6.65CGDD226 pKa = 4.61EE227 pKa = 5.87GSDD230 pKa = 3.1TWLDD234 pKa = 3.57VPGTQQIGTVAGSSFPTSDD253 pKa = 3.66SAWVEE258 pKa = 4.11VTGSSTVPTAEE269 pKa = 3.92VNDD272 pKa = 4.11YY273 pKa = 11.25EE274 pKa = 5.19GGTVTVTGSTEE285 pKa = 4.02APVGASVCRR294 pKa = 11.84SGQTTGWHH302 pKa = 6.12CGVIEE307 pKa = 5.26AKK309 pKa = 10.48DD310 pKa = 3.32QTVRR314 pKa = 11.84YY315 pKa = 9.23AGNLVVYY322 pKa = 10.27GLTRR326 pKa = 11.84TSACAEE332 pKa = 4.31GGDD335 pKa = 5.2SGGSWLAGTEE345 pKa = 4.19AQGVTSGGSGNCTFGGTTFFQPLNPILDD373 pKa = 3.82EE374 pKa = 4.24WDD376 pKa = 3.38LTLLTGG382 pKa = 4.58
Molecular weight: 39.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q9TFL8|A0A1Q9TFL8_9ACTN 50S ribosomal protein L30 OS=Nocardiopsis sp. CNR-923 OX=1904965 GN=rpmD PE=3 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.54GRR40 pKa = 11.84SALTVSHH47 pKa = 7.04
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.54GRR40 pKa = 11.84SALTVSHH47 pKa = 7.04
Molecular weight: 5.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1021308 |
37 |
2136 |
271.9 |
29.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.789 ± 0.047 | 0.82 ± 0.013 |
6.388 ± 0.037 | 6.337 ± 0.037 |
2.7 ± 0.023 | 9.196 ± 0.041 |
2.434 ± 0.022 | 3.089 ± 0.024 |
1.549 ± 0.026 | 10.172 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.885 ± 0.017 | 1.697 ± 0.02 |
5.971 ± 0.039 | 2.532 ± 0.019 |
8.858 ± 0.048 | 5.211 ± 0.031 |
5.872 ± 0.031 | 8.975 ± 0.043 |
1.588 ± 0.021 | 1.935 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
