
Clostridium aceticum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
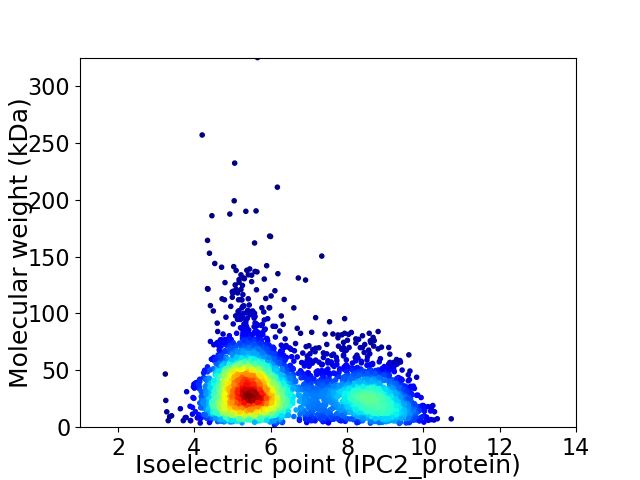
Virtual 2D-PAGE plot for 3835 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D8IBX6|A0A0D8IBX6_9CLOT Uncharacterized protein OS=Clostridium aceticum OX=84022 GN=CACET_c22820 PE=4 SV=1
MM1 pKa = 7.53SYY3 pKa = 10.54LRR5 pKa = 11.84KK6 pKa = 9.54RR7 pKa = 11.84LYY9 pKa = 10.34RR10 pKa = 11.84KK11 pKa = 9.84ISITVLLILSLVVMAACSPEE31 pKa = 4.03GTDD34 pKa = 3.85EE35 pKa = 5.34RR36 pKa = 11.84IVLADD41 pKa = 4.28AGWDD45 pKa = 3.14SLAFHH50 pKa = 6.85NEE52 pKa = 3.16VAALIIEE59 pKa = 4.25NGYY62 pKa = 10.69GYY64 pKa = 8.67STYY67 pKa = 10.23IEE69 pKa = 4.39MGSTPITFAALRR81 pKa = 11.84NGSIDD86 pKa = 3.2VYY88 pKa = 10.82MEE90 pKa = 4.39LWTDD94 pKa = 3.88NIIEE98 pKa = 4.95AYY100 pKa = 9.46TEE102 pKa = 3.93ALEE105 pKa = 4.37EE106 pKa = 4.17GDD108 pKa = 3.8IIEE111 pKa = 4.44VSTNFDD117 pKa = 3.79DD118 pKa = 4.34NAQGLYY124 pKa = 10.0VPTYY128 pKa = 10.12VIEE131 pKa = 4.77GDD133 pKa = 3.43AEE135 pKa = 4.16RR136 pKa = 11.84GIEE139 pKa = 3.99PMAPGLRR146 pKa = 11.84SVQDD150 pKa = 3.74LPDD153 pKa = 3.25YY154 pKa = 10.93WEE156 pKa = 4.14VFKK159 pKa = 11.3DD160 pKa = 3.88PEE162 pKa = 4.5DD163 pKa = 3.64SSKK166 pKa = 10.93GRR168 pKa = 11.84IYY170 pKa = 10.85GAIPGWEE177 pKa = 3.94VDD179 pKa = 4.4EE180 pKa = 5.3ILQQKK185 pKa = 9.3VKK187 pKa = 9.11THH189 pKa = 6.25GLDD192 pKa = 2.86EE193 pKa = 4.6NYY195 pKa = 10.85NYY197 pKa = 10.05FSPGSDD203 pKa = 2.9TALGSSIIAAIEE215 pKa = 3.99RR216 pKa = 11.84GEE218 pKa = 4.01AWLGYY223 pKa = 8.21YY224 pKa = 8.93WEE226 pKa = 4.43PTWIIGMYY234 pKa = 10.93DD235 pKa = 3.21MTLLEE240 pKa = 5.57DD241 pKa = 3.33IPYY244 pKa = 10.67DD245 pKa = 3.65EE246 pKa = 5.39EE247 pKa = 4.08KK248 pKa = 10.72WNDD251 pKa = 3.49GYY253 pKa = 10.42ATEE256 pKa = 4.45MPAVDD261 pKa = 3.5VTVAVHH267 pKa = 7.07KK268 pKa = 10.66DD269 pKa = 3.45MPEE272 pKa = 3.84KK273 pKa = 10.65APEE276 pKa = 3.61IVAFLEE282 pKa = 4.37NYY284 pKa = 6.14QTSSAVTSEE293 pKa = 3.66ALAYY297 pKa = 8.45MQANDD302 pKa = 3.5ATTEE306 pKa = 3.92EE307 pKa = 4.08AAVWFLKK314 pKa = 10.31EE315 pKa = 5.32YY316 pKa = 10.48EE317 pKa = 4.59DD318 pKa = 5.73LWTAWVPEE326 pKa = 4.28EE327 pKa = 3.64VAEE330 pKa = 4.11AVKK333 pKa = 10.33EE334 pKa = 4.22AIQQ337 pKa = 3.64
MM1 pKa = 7.53SYY3 pKa = 10.54LRR5 pKa = 11.84KK6 pKa = 9.54RR7 pKa = 11.84LYY9 pKa = 10.34RR10 pKa = 11.84KK11 pKa = 9.84ISITVLLILSLVVMAACSPEE31 pKa = 4.03GTDD34 pKa = 3.85EE35 pKa = 5.34RR36 pKa = 11.84IVLADD41 pKa = 4.28AGWDD45 pKa = 3.14SLAFHH50 pKa = 6.85NEE52 pKa = 3.16VAALIIEE59 pKa = 4.25NGYY62 pKa = 10.69GYY64 pKa = 8.67STYY67 pKa = 10.23IEE69 pKa = 4.39MGSTPITFAALRR81 pKa = 11.84NGSIDD86 pKa = 3.2VYY88 pKa = 10.82MEE90 pKa = 4.39LWTDD94 pKa = 3.88NIIEE98 pKa = 4.95AYY100 pKa = 9.46TEE102 pKa = 3.93ALEE105 pKa = 4.37EE106 pKa = 4.17GDD108 pKa = 3.8IIEE111 pKa = 4.44VSTNFDD117 pKa = 3.79DD118 pKa = 4.34NAQGLYY124 pKa = 10.0VPTYY128 pKa = 10.12VIEE131 pKa = 4.77GDD133 pKa = 3.43AEE135 pKa = 4.16RR136 pKa = 11.84GIEE139 pKa = 3.99PMAPGLRR146 pKa = 11.84SVQDD150 pKa = 3.74LPDD153 pKa = 3.25YY154 pKa = 10.93WEE156 pKa = 4.14VFKK159 pKa = 11.3DD160 pKa = 3.88PEE162 pKa = 4.5DD163 pKa = 3.64SSKK166 pKa = 10.93GRR168 pKa = 11.84IYY170 pKa = 10.85GAIPGWEE177 pKa = 3.94VDD179 pKa = 4.4EE180 pKa = 5.3ILQQKK185 pKa = 9.3VKK187 pKa = 9.11THH189 pKa = 6.25GLDD192 pKa = 2.86EE193 pKa = 4.6NYY195 pKa = 10.85NYY197 pKa = 10.05FSPGSDD203 pKa = 2.9TALGSSIIAAIEE215 pKa = 3.99RR216 pKa = 11.84GEE218 pKa = 4.01AWLGYY223 pKa = 8.21YY224 pKa = 8.93WEE226 pKa = 4.43PTWIIGMYY234 pKa = 10.93DD235 pKa = 3.21MTLLEE240 pKa = 5.57DD241 pKa = 3.33IPYY244 pKa = 10.67DD245 pKa = 3.65EE246 pKa = 5.39EE247 pKa = 4.08KK248 pKa = 10.72WNDD251 pKa = 3.49GYY253 pKa = 10.42ATEE256 pKa = 4.45MPAVDD261 pKa = 3.5VTVAVHH267 pKa = 7.07KK268 pKa = 10.66DD269 pKa = 3.45MPEE272 pKa = 3.84KK273 pKa = 10.65APEE276 pKa = 3.61IVAFLEE282 pKa = 4.37NYY284 pKa = 6.14QTSSAVTSEE293 pKa = 3.66ALAYY297 pKa = 8.45MQANDD302 pKa = 3.5ATTEE306 pKa = 3.92EE307 pKa = 4.08AAVWFLKK314 pKa = 10.31EE315 pKa = 5.32YY316 pKa = 10.48EE317 pKa = 4.59DD318 pKa = 5.73LWTAWVPEE326 pKa = 4.28EE327 pKa = 3.64VAEE330 pKa = 4.11AVKK333 pKa = 10.33EE334 pKa = 4.22AIQQ337 pKa = 3.64
Molecular weight: 37.86 kDa
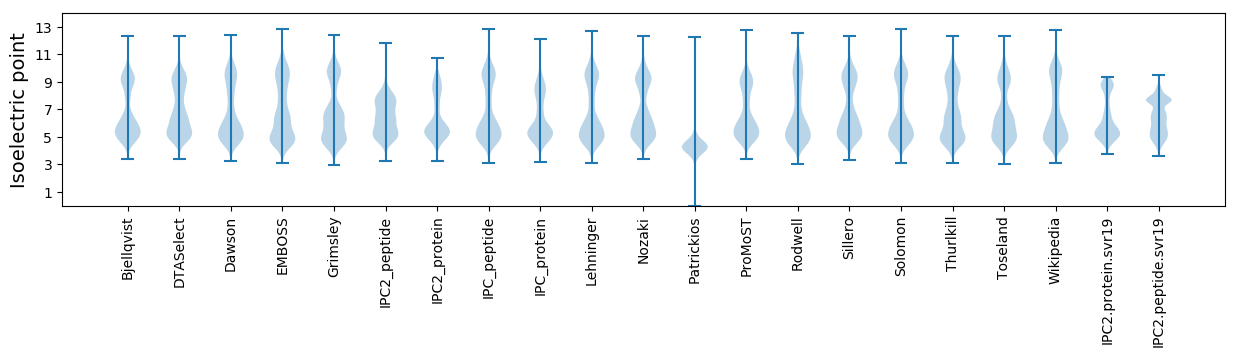
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D8IAJ2|A0A0D8IAJ2_9CLOT Uncharacterized protein OS=Clostridium aceticum OX=84022 GN=CACET_c25770 PE=4 SV=1
MM1 pKa = 7.34MGDD4 pKa = 3.26KK5 pKa = 11.05LNIIRR10 pKa = 11.84LGYY13 pKa = 9.91RR14 pKa = 11.84KK15 pKa = 9.52RR16 pKa = 11.84QIRR19 pKa = 11.84WIVVTLILATITLSLCVTLLLLGNSKK45 pKa = 10.84YY46 pKa = 10.66SLEE49 pKa = 4.1VVIRR53 pKa = 11.84VLLGEE58 pKa = 4.91EE59 pKa = 3.72IQGASFAISTLRR71 pKa = 11.84LPRR74 pKa = 11.84MLAGLLVGIGFGMAGSTFQTILRR97 pKa = 11.84NPLASPDD104 pKa = 3.96IIGVTAGSSVGAMFCILVLKK124 pKa = 10.23MSGTIVSVVSVISGLLVAVLIYY146 pKa = 10.72LFSKK150 pKa = 10.8VGGFSGGKK158 pKa = 9.8LILIGIGIQAMLNAVVSFLLLKK180 pKa = 10.52APQYY184 pKa = 10.29DD185 pKa = 3.46VPAAYY190 pKa = 9.41RR191 pKa = 11.84WLSGSLNGIQMTNIPSLFIVLVIFGSVIILLGRR224 pKa = 11.84HH225 pKa = 5.73LKK227 pKa = 10.1ILEE230 pKa = 4.64LGEE233 pKa = 4.16QSATTLGVRR242 pKa = 11.84ADD244 pKa = 3.8LVRR247 pKa = 11.84FLLVLSAVFLIAFATAVTGPISFVAFLAGPIATRR281 pKa = 11.84VVGSGAPNEE290 pKa = 4.3FPAGLVGAILVLGADD305 pKa = 5.37LIGQFAFGTRR315 pKa = 11.84FPVGVITGILGAPYY329 pKa = 10.48LIILLIRR336 pKa = 11.84MNRR339 pKa = 11.84TGGSAA344 pKa = 3.0
MM1 pKa = 7.34MGDD4 pKa = 3.26KK5 pKa = 11.05LNIIRR10 pKa = 11.84LGYY13 pKa = 9.91RR14 pKa = 11.84KK15 pKa = 9.52RR16 pKa = 11.84QIRR19 pKa = 11.84WIVVTLILATITLSLCVTLLLLGNSKK45 pKa = 10.84YY46 pKa = 10.66SLEE49 pKa = 4.1VVIRR53 pKa = 11.84VLLGEE58 pKa = 4.91EE59 pKa = 3.72IQGASFAISTLRR71 pKa = 11.84LPRR74 pKa = 11.84MLAGLLVGIGFGMAGSTFQTILRR97 pKa = 11.84NPLASPDD104 pKa = 3.96IIGVTAGSSVGAMFCILVLKK124 pKa = 10.23MSGTIVSVVSVISGLLVAVLIYY146 pKa = 10.72LFSKK150 pKa = 10.8VGGFSGGKK158 pKa = 9.8LILIGIGIQAMLNAVVSFLLLKK180 pKa = 10.52APQYY184 pKa = 10.29DD185 pKa = 3.46VPAAYY190 pKa = 9.41RR191 pKa = 11.84WLSGSLNGIQMTNIPSLFIVLVIFGSVIILLGRR224 pKa = 11.84HH225 pKa = 5.73LKK227 pKa = 10.1ILEE230 pKa = 4.64LGEE233 pKa = 4.16QSATTLGVRR242 pKa = 11.84ADD244 pKa = 3.8LVRR247 pKa = 11.84FLLVLSAVFLIAFATAVTGPISFVAFLAGPIATRR281 pKa = 11.84VVGSGAPNEE290 pKa = 4.3FPAGLVGAILVLGADD305 pKa = 5.37LIGQFAFGTRR315 pKa = 11.84FPVGVITGILGAPYY329 pKa = 10.48LIILLIRR336 pKa = 11.84MNRR339 pKa = 11.84TGGSAA344 pKa = 3.0
Molecular weight: 36.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1179242 |
14 |
2831 |
307.5 |
34.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.535 ± 0.046 | 0.99 ± 0.017 |
5.029 ± 0.034 | 7.851 ± 0.046 |
4.161 ± 0.03 | 6.774 ± 0.045 |
1.787 ± 0.016 | 9.243 ± 0.039 |
7.789 ± 0.048 | 9.589 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.849 ± 0.019 | 4.789 ± 0.026 |
3.21 ± 0.028 | 3.266 ± 0.02 |
3.802 ± 0.026 | 5.648 ± 0.03 |
5.27 ± 0.036 | 6.9 ± 0.037 |
0.768 ± 0.015 | 3.75 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
