
Aspergillus niger (strain CBS 513.88 / FGSC A1513)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Aspergillus subgen. Circumdati; Aspergillus niger
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
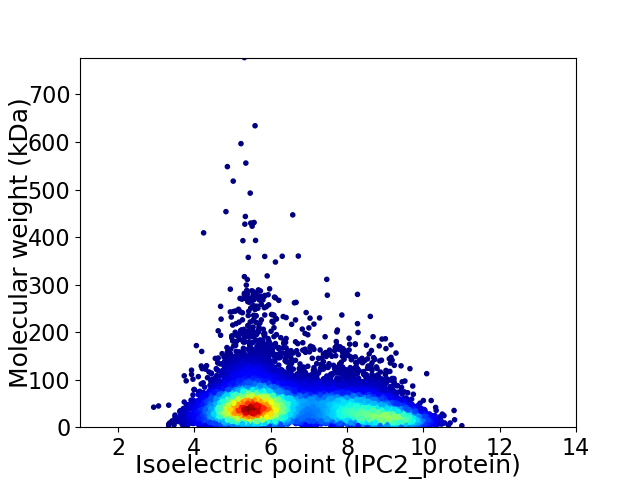
Virtual 2D-PAGE plot for 14068 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A2QH82|A2QH82_ASPNC Aspergillus niger contig An03c0180 genomic contig OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=An03g05930 PE=4 SV=1
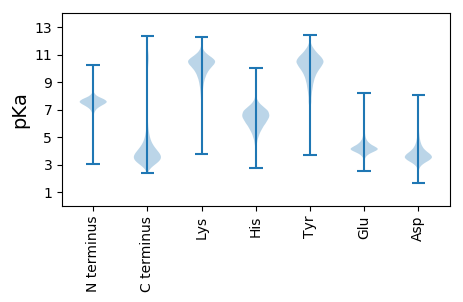
MM1 pKa = 7.59AFKK4 pKa = 8.7TTAGLAFLALAGSVKK19 pKa = 10.35AQSVDD24 pKa = 2.83GSKK27 pKa = 11.16YY28 pKa = 9.83NSPSNGPPASYY39 pKa = 9.75FAAATTLPVAALQSAAAKK57 pKa = 10.14ASSVPSKK64 pKa = 10.16ATYY67 pKa = 9.19PVNTDD72 pKa = 3.23DD73 pKa = 6.32DD74 pKa = 4.58SPKK77 pKa = 10.06STIHH81 pKa = 6.41SDD83 pKa = 2.72WVKK86 pKa = 10.45FNQGAALSWVADD98 pKa = 3.6MDD100 pKa = 4.13VDD102 pKa = 5.35CDD104 pKa = 4.89GIDD107 pKa = 3.7YY108 pKa = 10.19KK109 pKa = 11.38CKK111 pKa = 10.76GNGDD115 pKa = 4.15GLPEE119 pKa = 4.35TNWGALSAYY128 pKa = 6.81EE129 pKa = 4.22VPWIVIPDD137 pKa = 3.55QFLTANEE144 pKa = 4.25DD145 pKa = 3.83LLPGNNVAAVICNGKK160 pKa = 8.75MYY162 pKa = 10.7YY163 pKa = 10.53GILGDD168 pKa = 4.46SNGDD172 pKa = 3.56DD173 pKa = 3.61PEE175 pKa = 4.56VTGEE179 pKa = 4.26ASWLMARR186 pKa = 11.84TCFPDD191 pKa = 4.84DD192 pKa = 4.5DD193 pKa = 5.37LNGAEE198 pKa = 4.32GHH200 pKa = 6.72AEE202 pKa = 3.82ADD204 pKa = 4.28VTCKK208 pKa = 10.36PYY210 pKa = 10.24TRR212 pKa = 11.84SALNKK217 pKa = 10.09NYY219 pKa = 8.33ITNFSTLRR227 pKa = 11.84SMGDD231 pKa = 3.28KK232 pKa = 10.77LVGALASNLGLTSSASGSTATCSWQGHH259 pKa = 5.4CEE261 pKa = 3.74GAICSTEE268 pKa = 4.19DD269 pKa = 3.48DD270 pKa = 4.32CSDD273 pKa = 5.38DD274 pKa = 4.41LVCDD278 pKa = 3.72SGKK281 pKa = 10.45CSSPDD286 pKa = 3.29EE287 pKa = 5.13DD288 pKa = 6.05DD289 pKa = 6.01GDD291 pKa = 5.31DD292 pKa = 4.57EE293 pKa = 6.49DD294 pKa = 6.8DD295 pKa = 4.03EE296 pKa = 6.29DD297 pKa = 5.26EE298 pKa = 4.72EE299 pKa = 5.4EE300 pKa = 4.42NDD302 pKa = 5.03EE303 pKa = 5.64DD304 pKa = 6.44DD305 pKa = 5.6EE306 pKa = 7.61DD307 pKa = 6.7DD308 pKa = 5.74DD309 pKa = 7.19DD310 pKa = 6.53EE311 pKa = 7.71DD312 pKa = 6.68DD313 pKa = 5.05EE314 pKa = 7.16DD315 pKa = 6.26DD316 pKa = 3.82EE317 pKa = 4.83
MM1 pKa = 7.59AFKK4 pKa = 8.7TTAGLAFLALAGSVKK19 pKa = 10.35AQSVDD24 pKa = 2.83GSKK27 pKa = 11.16YY28 pKa = 9.83NSPSNGPPASYY39 pKa = 9.75FAAATTLPVAALQSAAAKK57 pKa = 10.14ASSVPSKK64 pKa = 10.16ATYY67 pKa = 9.19PVNTDD72 pKa = 3.23DD73 pKa = 6.32DD74 pKa = 4.58SPKK77 pKa = 10.06STIHH81 pKa = 6.41SDD83 pKa = 2.72WVKK86 pKa = 10.45FNQGAALSWVADD98 pKa = 3.6MDD100 pKa = 4.13VDD102 pKa = 5.35CDD104 pKa = 4.89GIDD107 pKa = 3.7YY108 pKa = 10.19KK109 pKa = 11.38CKK111 pKa = 10.76GNGDD115 pKa = 4.15GLPEE119 pKa = 4.35TNWGALSAYY128 pKa = 6.81EE129 pKa = 4.22VPWIVIPDD137 pKa = 3.55QFLTANEE144 pKa = 4.25DD145 pKa = 3.83LLPGNNVAAVICNGKK160 pKa = 8.75MYY162 pKa = 10.7YY163 pKa = 10.53GILGDD168 pKa = 4.46SNGDD172 pKa = 3.56DD173 pKa = 3.61PEE175 pKa = 4.56VTGEE179 pKa = 4.26ASWLMARR186 pKa = 11.84TCFPDD191 pKa = 4.84DD192 pKa = 4.5DD193 pKa = 5.37LNGAEE198 pKa = 4.32GHH200 pKa = 6.72AEE202 pKa = 3.82ADD204 pKa = 4.28VTCKK208 pKa = 10.36PYY210 pKa = 10.24TRR212 pKa = 11.84SALNKK217 pKa = 10.09NYY219 pKa = 8.33ITNFSTLRR227 pKa = 11.84SMGDD231 pKa = 3.28KK232 pKa = 10.77LVGALASNLGLTSSASGSTATCSWQGHH259 pKa = 5.4CEE261 pKa = 3.74GAICSTEE268 pKa = 4.19DD269 pKa = 3.48DD270 pKa = 4.32CSDD273 pKa = 5.38DD274 pKa = 4.41LVCDD278 pKa = 3.72SGKK281 pKa = 10.45CSSPDD286 pKa = 3.29EE287 pKa = 5.13DD288 pKa = 6.05DD289 pKa = 6.01GDD291 pKa = 5.31DD292 pKa = 4.57EE293 pKa = 6.49DD294 pKa = 6.8DD295 pKa = 4.03EE296 pKa = 6.29DD297 pKa = 5.26EE298 pKa = 4.72EE299 pKa = 5.4EE300 pKa = 4.42NDD302 pKa = 5.03EE303 pKa = 5.64DD304 pKa = 6.44DD305 pKa = 5.6EE306 pKa = 7.61DD307 pKa = 6.7DD308 pKa = 5.74DD309 pKa = 7.19DD310 pKa = 6.53EE311 pKa = 7.71DD312 pKa = 6.68DD313 pKa = 5.05EE314 pKa = 7.16DD315 pKa = 6.26DD316 pKa = 3.82EE317 pKa = 4.83
Molecular weight: 33.43 kDa
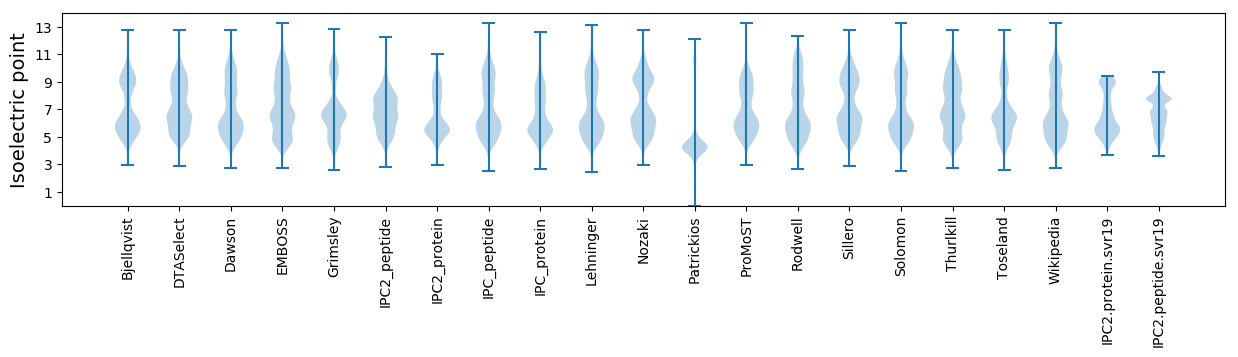
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A2RA46|A2RA46_ASPNC Aspergillus niger contig An18c0050 genomic contig OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=An18g01890 PE=4 SV=1
MM1 pKa = 7.04VNRR4 pKa = 11.84RR5 pKa = 11.84LRR7 pKa = 11.84VRR9 pKa = 11.84IRR11 pKa = 11.84ISGRR15 pKa = 11.84AVLRR19 pKa = 11.84TNIFNKK25 pKa = 9.78VV26 pKa = 2.87
MM1 pKa = 7.04VNRR4 pKa = 11.84RR5 pKa = 11.84LRR7 pKa = 11.84VRR9 pKa = 11.84IRR11 pKa = 11.84ISGRR15 pKa = 11.84AVLRR19 pKa = 11.84TNIFNKK25 pKa = 9.78VV26 pKa = 2.87
Molecular weight: 3.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6183668 |
10 |
7035 |
439.6 |
48.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.363 ± 0.018 | 1.411 ± 0.009 |
5.506 ± 0.014 | 5.987 ± 0.024 |
3.656 ± 0.012 | 6.859 ± 0.018 |
2.513 ± 0.01 | 4.985 ± 0.015 |
4.386 ± 0.02 | 9.155 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.209 ± 0.008 | 3.571 ± 0.01 |
6.069 ± 0.021 | 4.011 ± 0.014 |
6.259 ± 0.021 | 8.391 ± 0.023 |
6.017 ± 0.015 | 6.208 ± 0.015 |
1.524 ± 0.008 | 2.916 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
