
Halapricum salinum
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Halapricum
Average proteome isoelectric point is 4.83
Get precalculated fractions of proteins
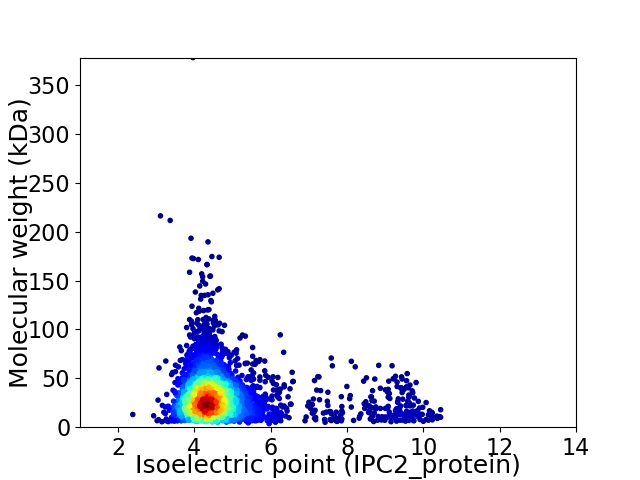
Virtual 2D-PAGE plot for 3303 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
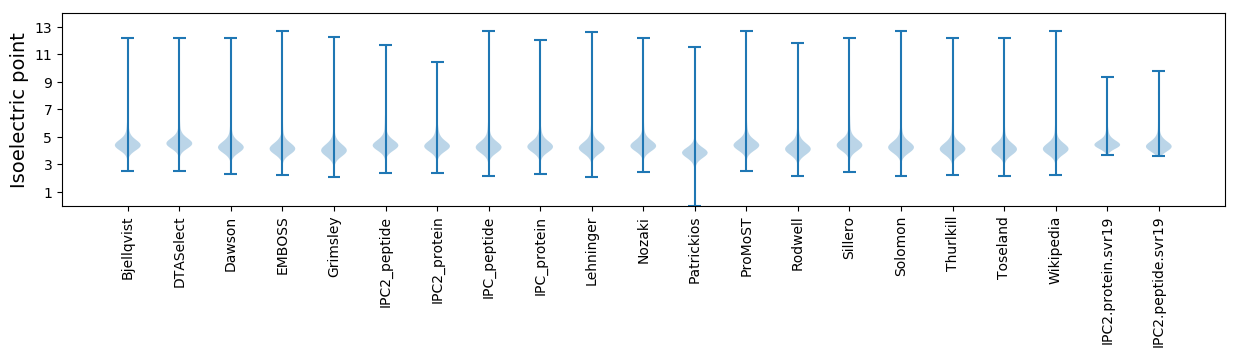
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6HIR9|A0A4D6HIR9_9EURY 2 5-diamino-6-(5-phospho-D-ribosylamino)pyrimidin-4(3H)-one reductase OS=Halapricum salinum OX=1457250 GN=DV733_15775 PE=3 SV=1
MM1 pKa = 7.95PLTPGDD7 pKa = 4.1DD8 pKa = 3.82APTFAAQNQHH18 pKa = 6.25DD19 pKa = 4.25KK20 pKa = 11.14RR21 pKa = 11.84VVCQYY26 pKa = 11.08DD27 pKa = 3.62RR28 pKa = 11.84PTVVYY33 pKa = 7.9FYY35 pKa = 10.53PRR37 pKa = 11.84DD38 pKa = 3.79DD39 pKa = 3.98TPGCTIEE46 pKa = 5.8AEE48 pKa = 4.3GFDD51 pKa = 4.9DD52 pKa = 4.38SLSAYY57 pKa = 9.92RR58 pKa = 11.84EE59 pKa = 4.17AGVTVYY65 pKa = 10.47GVSTDD70 pKa = 3.33DD71 pKa = 3.83VEE73 pKa = 4.52SHH75 pKa = 7.01RR76 pKa = 11.84EE77 pKa = 3.58FADD80 pKa = 3.89EE81 pKa = 3.86YY82 pKa = 11.41DD83 pKa = 3.29IAFDD87 pKa = 5.36LLADD91 pKa = 4.22PDD93 pKa = 4.33GNIADD98 pKa = 4.09SFDD101 pKa = 3.61VPVEE105 pKa = 3.87NGAAARR111 pKa = 11.84TTFVVVDD118 pKa = 4.03GQVVATYY125 pKa = 10.43EE126 pKa = 4.36GVHH129 pKa = 6.47PEE131 pKa = 3.57GHH133 pKa = 6.33AADD136 pKa = 3.73VLEE139 pKa = 4.97DD140 pKa = 3.72LVEE143 pKa = 4.16TGIVSPP149 pKa = 4.86
MM1 pKa = 7.95PLTPGDD7 pKa = 4.1DD8 pKa = 3.82APTFAAQNQHH18 pKa = 6.25DD19 pKa = 4.25KK20 pKa = 11.14RR21 pKa = 11.84VVCQYY26 pKa = 11.08DD27 pKa = 3.62RR28 pKa = 11.84PTVVYY33 pKa = 7.9FYY35 pKa = 10.53PRR37 pKa = 11.84DD38 pKa = 3.79DD39 pKa = 3.98TPGCTIEE46 pKa = 5.8AEE48 pKa = 4.3GFDD51 pKa = 4.9DD52 pKa = 4.38SLSAYY57 pKa = 9.92RR58 pKa = 11.84EE59 pKa = 4.17AGVTVYY65 pKa = 10.47GVSTDD70 pKa = 3.33DD71 pKa = 3.83VEE73 pKa = 4.52SHH75 pKa = 7.01RR76 pKa = 11.84EE77 pKa = 3.58FADD80 pKa = 3.89EE81 pKa = 3.86YY82 pKa = 11.41DD83 pKa = 3.29IAFDD87 pKa = 5.36LLADD91 pKa = 4.22PDD93 pKa = 4.33GNIADD98 pKa = 4.09SFDD101 pKa = 3.61VPVEE105 pKa = 3.87NGAAARR111 pKa = 11.84TTFVVVDD118 pKa = 4.03GQVVATYY125 pKa = 10.43EE126 pKa = 4.36GVHH129 pKa = 6.47PEE131 pKa = 3.57GHH133 pKa = 6.33AADD136 pKa = 3.73VLEE139 pKa = 4.97DD140 pKa = 3.72LVEE143 pKa = 4.16TGIVSPP149 pKa = 4.86
Molecular weight: 16.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6HBX2|A0A4D6HBX2_9EURY Ribose-5-phosphate isomerase A OS=Halapricum salinum OX=1457250 GN=rpiA PE=3 SV=1
MM1 pKa = 6.88YY2 pKa = 10.32LRR4 pKa = 11.84RR5 pKa = 11.84IEE7 pKa = 4.22EE8 pKa = 4.31LPLEE12 pKa = 4.7DD13 pKa = 5.6RR14 pKa = 11.84LDD16 pKa = 3.8APADD20 pKa = 3.83RR21 pKa = 11.84KK22 pKa = 10.5ADD24 pKa = 3.88RR25 pKa = 11.84EE26 pKa = 4.6TPFLLGTTFSASGFLARR43 pKa = 11.84FVEE46 pKa = 4.86RR47 pKa = 11.84AAGTALRR54 pKa = 11.84LRR56 pKa = 11.84RR57 pKa = 11.84NGIRR61 pKa = 11.84SRR63 pKa = 11.84ASPATRR69 pKa = 11.84GAKK72 pKa = 8.27TWGKK76 pKa = 10.17KK77 pKa = 9.47PEE79 pKa = 4.36SPWEE83 pKa = 4.21GDD85 pKa = 3.34SGEE88 pKa = 4.29TATLSPYY95 pKa = 10.66AGQLYY100 pKa = 9.84SSRR103 pKa = 11.84PALPRR108 pKa = 11.84VARR111 pKa = 11.84SRR113 pKa = 11.84VTAFPVRR120 pKa = 11.84NRR122 pKa = 11.84DD123 pKa = 3.47APCGRR128 pKa = 11.84VSHH131 pKa = 7.01ALPATAGSRR140 pKa = 11.84TCRR143 pKa = 11.84GHH145 pKa = 5.74TLASGVTFKK154 pKa = 10.96SLRR157 pKa = 11.84SPVRR161 pKa = 11.84DD162 pKa = 3.39VSEE165 pKa = 4.54TPSPAPFLPRR175 pKa = 11.84TAAFLCDD182 pKa = 3.54QVVVLVLVVGPLAVAGVDD200 pKa = 3.38VLAPANRR207 pKa = 11.84TPIFLALMGAAFTYY221 pKa = 10.49HH222 pKa = 7.04FLLEE226 pKa = 4.25WLFGTTLGKK235 pKa = 10.31RR236 pKa = 11.84GFDD239 pKa = 3.46LRR241 pKa = 11.84VVADD245 pKa = 4.34DD246 pKa = 4.27GQPLGLWGSFVRR258 pKa = 11.84NALRR262 pKa = 11.84LIDD265 pKa = 4.32GLGYY269 pKa = 7.86WTVATVILFYY279 pKa = 10.91RR280 pKa = 11.84GDD282 pKa = 3.79GKK284 pKa = 11.12RR285 pKa = 11.84LGDD288 pKa = 3.46VLGRR292 pKa = 11.84TLVVRR297 pKa = 11.84ATTDD301 pKa = 2.75ARR303 pKa = 3.77
MM1 pKa = 6.88YY2 pKa = 10.32LRR4 pKa = 11.84RR5 pKa = 11.84IEE7 pKa = 4.22EE8 pKa = 4.31LPLEE12 pKa = 4.7DD13 pKa = 5.6RR14 pKa = 11.84LDD16 pKa = 3.8APADD20 pKa = 3.83RR21 pKa = 11.84KK22 pKa = 10.5ADD24 pKa = 3.88RR25 pKa = 11.84EE26 pKa = 4.6TPFLLGTTFSASGFLARR43 pKa = 11.84FVEE46 pKa = 4.86RR47 pKa = 11.84AAGTALRR54 pKa = 11.84LRR56 pKa = 11.84RR57 pKa = 11.84NGIRR61 pKa = 11.84SRR63 pKa = 11.84ASPATRR69 pKa = 11.84GAKK72 pKa = 8.27TWGKK76 pKa = 10.17KK77 pKa = 9.47PEE79 pKa = 4.36SPWEE83 pKa = 4.21GDD85 pKa = 3.34SGEE88 pKa = 4.29TATLSPYY95 pKa = 10.66AGQLYY100 pKa = 9.84SSRR103 pKa = 11.84PALPRR108 pKa = 11.84VARR111 pKa = 11.84SRR113 pKa = 11.84VTAFPVRR120 pKa = 11.84NRR122 pKa = 11.84DD123 pKa = 3.47APCGRR128 pKa = 11.84VSHH131 pKa = 7.01ALPATAGSRR140 pKa = 11.84TCRR143 pKa = 11.84GHH145 pKa = 5.74TLASGVTFKK154 pKa = 10.96SLRR157 pKa = 11.84SPVRR161 pKa = 11.84DD162 pKa = 3.39VSEE165 pKa = 4.54TPSPAPFLPRR175 pKa = 11.84TAAFLCDD182 pKa = 3.54QVVVLVLVVGPLAVAGVDD200 pKa = 3.38VLAPANRR207 pKa = 11.84TPIFLALMGAAFTYY221 pKa = 10.49HH222 pKa = 7.04FLLEE226 pKa = 4.25WLFGTTLGKK235 pKa = 10.31RR236 pKa = 11.84GFDD239 pKa = 3.46LRR241 pKa = 11.84VVADD245 pKa = 4.34DD246 pKa = 4.27GQPLGLWGSFVRR258 pKa = 11.84NALRR262 pKa = 11.84LIDD265 pKa = 4.32GLGYY269 pKa = 7.86WTVATVILFYY279 pKa = 10.91RR280 pKa = 11.84GDD282 pKa = 3.79GKK284 pKa = 11.12RR285 pKa = 11.84LGDD288 pKa = 3.46VLGRR292 pKa = 11.84TLVVRR297 pKa = 11.84ATTDD301 pKa = 2.75ARR303 pKa = 3.77
Molecular weight: 32.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1007341 |
30 |
3572 |
305.0 |
33.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.407 ± 0.062 | 0.695 ± 0.012 |
8.359 ± 0.054 | 8.614 ± 0.065 |
3.324 ± 0.025 | 8.411 ± 0.047 |
1.988 ± 0.021 | 4.288 ± 0.03 |
1.847 ± 0.026 | 8.974 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.714 ± 0.016 | 2.355 ± 0.034 |
4.616 ± 0.028 | 2.904 ± 0.026 |
6.397 ± 0.043 | 5.7 ± 0.041 |
6.542 ± 0.043 | 8.922 ± 0.051 |
1.191 ± 0.018 | 2.753 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
