
Changjiang narna-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KI63|A0A1L3KI63_9VIRU RNA-dependent RNA polymerase OS=Changjiang narna-like virus 1 OX=1922776 PE=4 SV=1
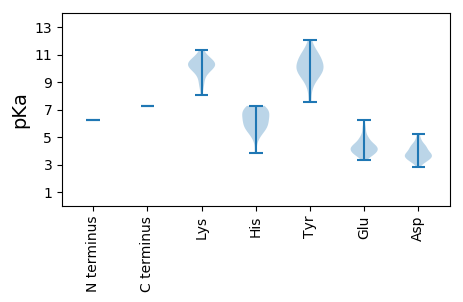
MM1 pKa = 6.24MTEE4 pKa = 4.95SEE6 pKa = 4.69PLMCPDD12 pKa = 4.2SSSLSAPIGFAAALPKK28 pKa = 10.16KK29 pKa = 9.86AAAVLTIEE37 pKa = 5.42NIEE40 pKa = 3.74SWRR43 pKa = 11.84RR44 pKa = 11.84LTSASLPTAGAPSWHH59 pKa = 6.99PLVVVDD65 pKa = 4.38SQQEE69 pKa = 4.1ILSHH73 pKa = 6.81SEE75 pKa = 3.76ALGPPLDD82 pKa = 5.21LPLIDD87 pKa = 4.98CFIKK91 pKa = 10.4ASHH94 pKa = 6.48MIHH97 pKa = 7.04WYY99 pKa = 10.38NNTVEE104 pKa = 4.21IWAHH108 pKa = 4.88SLISGDD114 pKa = 3.66YY115 pKa = 11.09GEE117 pKa = 5.02SLVNKK122 pKa = 9.7GCILRR127 pKa = 11.84SCLSTPTCSRR137 pKa = 11.84MLGRR141 pKa = 11.84WTALAQFGKK150 pKa = 10.82LEE152 pKa = 4.7AYY154 pKa = 9.61LKK156 pKa = 10.1WVTANFFAEE165 pKa = 4.18SLLQKK170 pKa = 9.71EE171 pKa = 5.01LPPPPDD177 pKa = 3.15VFKK180 pKa = 11.33GILAKK185 pKa = 10.28IPGMKK190 pKa = 9.43RR191 pKa = 11.84PYY193 pKa = 9.69PGRR196 pKa = 11.84SWSSLFSRR204 pKa = 11.84THH206 pKa = 5.05SVKK209 pKa = 10.77SGLKK213 pKa = 8.72WRR215 pKa = 11.84FTFGKK220 pKa = 9.95DD221 pKa = 3.43FYY223 pKa = 8.44MTKK226 pKa = 10.2NASLPVEE233 pKa = 4.12EE234 pKa = 5.69SFVQANLEE242 pKa = 3.97KK243 pKa = 10.33HH244 pKa = 5.63QKK246 pKa = 8.96ILCGQIEE253 pKa = 4.67DD254 pKa = 4.29KK255 pKa = 11.06LSEE258 pKa = 3.87ATYY261 pKa = 11.12DD262 pKa = 3.87EE263 pKa = 4.44VCEE266 pKa = 4.88AIQLCADD273 pKa = 4.5EE274 pKa = 5.6IFGKK278 pKa = 9.85PYY280 pKa = 9.97EE281 pKa = 4.33VPSKK285 pKa = 10.6FISSDD290 pKa = 3.62LEE292 pKa = 3.57ITEE295 pKa = 4.61NIRR298 pKa = 11.84SFSRR302 pKa = 11.84PEE304 pKa = 3.57IKK306 pKa = 10.35CPSRR310 pKa = 11.84LPSFGASFHH319 pKa = 6.34GNRR322 pKa = 11.84GDD324 pKa = 4.37GGACGDD330 pKa = 4.29LLRR333 pKa = 11.84NYY335 pKa = 10.01HH336 pKa = 7.26DD337 pKa = 4.05IGSLPEE343 pKa = 4.59PNDD346 pKa = 3.95GYY348 pKa = 11.46LWGYY352 pKa = 7.56AQKK355 pKa = 10.73NGSEE359 pKa = 4.12LCEE362 pKa = 3.87VRR364 pKa = 11.84TLHH367 pKa = 7.27DD368 pKa = 3.82PEE370 pKa = 5.26LFFEE374 pKa = 5.83AEE376 pKa = 4.37KK377 pKa = 10.33QWSRR381 pKa = 11.84EE382 pKa = 3.84ALFLSSEE389 pKa = 4.28GVNAHH394 pKa = 5.43VVPLIEE400 pKa = 4.24PFKK403 pKa = 11.16VRR405 pKa = 11.84TITKK409 pKa = 8.21GQAEE413 pKa = 4.76IYY415 pKa = 9.77HH416 pKa = 6.99LARR419 pKa = 11.84RR420 pKa = 11.84WQKK423 pKa = 10.27IIHH426 pKa = 5.65SRR428 pKa = 11.84MRR430 pKa = 11.84QHH432 pKa = 7.2PNFKK436 pKa = 10.75LIGQPCNGAFISQIFGNSKK455 pKa = 9.95LFNYY459 pKa = 10.44YY460 pKa = 10.24GDD462 pKa = 3.6KK463 pKa = 11.21NGFFVSGDD471 pKa = 3.67YY472 pKa = 11.0EE473 pKa = 4.48SATDD477 pKa = 4.34LLNPALSEE485 pKa = 4.03FAQEE489 pKa = 4.06QISIRR494 pKa = 11.84LGIPLEE500 pKa = 4.15DD501 pKa = 3.32QYY503 pKa = 12.06VLKK506 pKa = 10.76QCLTGHH512 pKa = 6.49RR513 pKa = 11.84LRR515 pKa = 11.84YY516 pKa = 9.15EE517 pKa = 3.85KK518 pKa = 10.54EE519 pKa = 3.95GQLFEE524 pKa = 4.32QTWGQLMGSPTSFPVLCLVNMAATLLSYY552 pKa = 10.85NRR554 pKa = 11.84AYY556 pKa = 9.94KK557 pKa = 10.56SSFRR561 pKa = 11.84LSDD564 pKa = 4.32LPTCVNGDD572 pKa = 4.0DD573 pKa = 3.69VLFWARR579 pKa = 11.84DD580 pKa = 3.43GAHH583 pKa = 5.54YY584 pKa = 9.02EE585 pKa = 3.33IWKK588 pKa = 10.19QITGEE593 pKa = 4.2CGLKK597 pKa = 10.43FSLGKK602 pKa = 10.35NYY604 pKa = 9.4TSRR607 pKa = 11.84HH608 pKa = 3.86VCVINSEE615 pKa = 4.34LYY617 pKa = 10.7RR618 pKa = 11.84FVKK621 pKa = 10.01DD622 pKa = 3.19TDD624 pKa = 4.1CQIHH628 pKa = 6.19RR629 pKa = 11.84PSPLFRR635 pKa = 11.84LEE637 pKa = 3.76KK638 pKa = 10.52ALNSRR643 pKa = 11.84LLCGGTRR650 pKa = 11.84SAASSGFDD658 pKa = 3.54PLMLSDD664 pKa = 4.7IDD666 pKa = 3.88LSIYY670 pKa = 10.42SSAVGGSAAFRR681 pKa = 11.84RR682 pKa = 11.84LVKK685 pKa = 10.48SHH687 pKa = 6.77LRR689 pKa = 11.84PHH691 pKa = 6.17QGVEE695 pKa = 3.84PSPKK699 pKa = 9.72KK700 pKa = 10.69VLAALRR706 pKa = 11.84KK707 pKa = 8.82RR708 pKa = 11.84YY709 pKa = 9.96RR710 pKa = 11.84NGGFSEE716 pKa = 6.23DD717 pKa = 3.44YY718 pKa = 10.83AKK720 pKa = 10.29WFNTIPSRR728 pKa = 11.84ALGLLEE734 pKa = 3.81QHH736 pKa = 6.91DD737 pKa = 4.74GEE739 pKa = 4.85IEE741 pKa = 3.82TDD743 pKa = 3.57LVVRR747 pKa = 11.84PKK749 pKa = 9.94MRR751 pKa = 11.84EE752 pKa = 3.55FMTKK756 pKa = 9.24VFNDD760 pKa = 3.51LQISKK765 pKa = 9.38LHH767 pKa = 5.94KK768 pKa = 9.69FARR771 pKa = 11.84ADD773 pKa = 3.35SRR775 pKa = 11.84VVKK778 pKa = 10.56NSPGFYY784 pKa = 10.09KK785 pKa = 10.15PQHH788 pKa = 6.17LGGLGLIPPINHH800 pKa = 5.41VFSIHH805 pKa = 6.51DD806 pKa = 3.7HH807 pKa = 6.96LEE809 pKa = 4.06IASLRR814 pKa = 11.84ALPAEE819 pKa = 3.95AHH821 pKa = 5.91QFSQGMTPKK830 pKa = 8.72MASVSFMEE838 pKa = 4.35SVRR841 pKa = 11.84AEE843 pKa = 3.68ILEE846 pKa = 4.23IQDD849 pKa = 3.32TLEE852 pKa = 3.94IEE854 pKa = 4.47RR855 pKa = 11.84ALLPAADD862 pKa = 3.72IEE864 pKa = 4.49SLRR867 pKa = 11.84FNGEE871 pKa = 3.63EE872 pKa = 3.99SQFWDD877 pKa = 3.2QEE879 pKa = 4.3FLVGFVDD886 pKa = 4.0PEE888 pKa = 4.17NVVVDD893 pKa = 3.69SEE895 pKa = 4.42SRR897 pKa = 11.84SRR899 pKa = 11.84KK900 pKa = 9.72EE901 pKa = 3.37SDD903 pKa = 3.15LNKK906 pKa = 10.16ILTSRR911 pKa = 11.84NWKK914 pKa = 9.25SRR916 pKa = 11.84RR917 pKa = 11.84LTGEE921 pKa = 3.64MRR923 pKa = 11.84RR924 pKa = 11.84QAALLEE930 pKa = 4.36KK931 pKa = 10.67ASGVKK936 pKa = 10.1CEE938 pKa = 4.27KK939 pKa = 10.17TFTDD943 pKa = 3.69ASHH946 pKa = 5.78GHH948 pKa = 5.85RR949 pKa = 11.84LRR951 pKa = 11.84YY952 pKa = 9.4LGAMPQGKK960 pKa = 8.05EE961 pKa = 3.94TVNLIEE967 pKa = 5.02GDD969 pKa = 2.82WHH971 pKa = 6.53LISRR975 pKa = 11.84RR976 pKa = 11.84PLPKK980 pKa = 10.12FHH982 pKa = 7.25
MM1 pKa = 6.24MTEE4 pKa = 4.95SEE6 pKa = 4.69PLMCPDD12 pKa = 4.2SSSLSAPIGFAAALPKK28 pKa = 10.16KK29 pKa = 9.86AAAVLTIEE37 pKa = 5.42NIEE40 pKa = 3.74SWRR43 pKa = 11.84RR44 pKa = 11.84LTSASLPTAGAPSWHH59 pKa = 6.99PLVVVDD65 pKa = 4.38SQQEE69 pKa = 4.1ILSHH73 pKa = 6.81SEE75 pKa = 3.76ALGPPLDD82 pKa = 5.21LPLIDD87 pKa = 4.98CFIKK91 pKa = 10.4ASHH94 pKa = 6.48MIHH97 pKa = 7.04WYY99 pKa = 10.38NNTVEE104 pKa = 4.21IWAHH108 pKa = 4.88SLISGDD114 pKa = 3.66YY115 pKa = 11.09GEE117 pKa = 5.02SLVNKK122 pKa = 9.7GCILRR127 pKa = 11.84SCLSTPTCSRR137 pKa = 11.84MLGRR141 pKa = 11.84WTALAQFGKK150 pKa = 10.82LEE152 pKa = 4.7AYY154 pKa = 9.61LKK156 pKa = 10.1WVTANFFAEE165 pKa = 4.18SLLQKK170 pKa = 9.71EE171 pKa = 5.01LPPPPDD177 pKa = 3.15VFKK180 pKa = 11.33GILAKK185 pKa = 10.28IPGMKK190 pKa = 9.43RR191 pKa = 11.84PYY193 pKa = 9.69PGRR196 pKa = 11.84SWSSLFSRR204 pKa = 11.84THH206 pKa = 5.05SVKK209 pKa = 10.77SGLKK213 pKa = 8.72WRR215 pKa = 11.84FTFGKK220 pKa = 9.95DD221 pKa = 3.43FYY223 pKa = 8.44MTKK226 pKa = 10.2NASLPVEE233 pKa = 4.12EE234 pKa = 5.69SFVQANLEE242 pKa = 3.97KK243 pKa = 10.33HH244 pKa = 5.63QKK246 pKa = 8.96ILCGQIEE253 pKa = 4.67DD254 pKa = 4.29KK255 pKa = 11.06LSEE258 pKa = 3.87ATYY261 pKa = 11.12DD262 pKa = 3.87EE263 pKa = 4.44VCEE266 pKa = 4.88AIQLCADD273 pKa = 4.5EE274 pKa = 5.6IFGKK278 pKa = 9.85PYY280 pKa = 9.97EE281 pKa = 4.33VPSKK285 pKa = 10.6FISSDD290 pKa = 3.62LEE292 pKa = 3.57ITEE295 pKa = 4.61NIRR298 pKa = 11.84SFSRR302 pKa = 11.84PEE304 pKa = 3.57IKK306 pKa = 10.35CPSRR310 pKa = 11.84LPSFGASFHH319 pKa = 6.34GNRR322 pKa = 11.84GDD324 pKa = 4.37GGACGDD330 pKa = 4.29LLRR333 pKa = 11.84NYY335 pKa = 10.01HH336 pKa = 7.26DD337 pKa = 4.05IGSLPEE343 pKa = 4.59PNDD346 pKa = 3.95GYY348 pKa = 11.46LWGYY352 pKa = 7.56AQKK355 pKa = 10.73NGSEE359 pKa = 4.12LCEE362 pKa = 3.87VRR364 pKa = 11.84TLHH367 pKa = 7.27DD368 pKa = 3.82PEE370 pKa = 5.26LFFEE374 pKa = 5.83AEE376 pKa = 4.37KK377 pKa = 10.33QWSRR381 pKa = 11.84EE382 pKa = 3.84ALFLSSEE389 pKa = 4.28GVNAHH394 pKa = 5.43VVPLIEE400 pKa = 4.24PFKK403 pKa = 11.16VRR405 pKa = 11.84TITKK409 pKa = 8.21GQAEE413 pKa = 4.76IYY415 pKa = 9.77HH416 pKa = 6.99LARR419 pKa = 11.84RR420 pKa = 11.84WQKK423 pKa = 10.27IIHH426 pKa = 5.65SRR428 pKa = 11.84MRR430 pKa = 11.84QHH432 pKa = 7.2PNFKK436 pKa = 10.75LIGQPCNGAFISQIFGNSKK455 pKa = 9.95LFNYY459 pKa = 10.44YY460 pKa = 10.24GDD462 pKa = 3.6KK463 pKa = 11.21NGFFVSGDD471 pKa = 3.67YY472 pKa = 11.0EE473 pKa = 4.48SATDD477 pKa = 4.34LLNPALSEE485 pKa = 4.03FAQEE489 pKa = 4.06QISIRR494 pKa = 11.84LGIPLEE500 pKa = 4.15DD501 pKa = 3.32QYY503 pKa = 12.06VLKK506 pKa = 10.76QCLTGHH512 pKa = 6.49RR513 pKa = 11.84LRR515 pKa = 11.84YY516 pKa = 9.15EE517 pKa = 3.85KK518 pKa = 10.54EE519 pKa = 3.95GQLFEE524 pKa = 4.32QTWGQLMGSPTSFPVLCLVNMAATLLSYY552 pKa = 10.85NRR554 pKa = 11.84AYY556 pKa = 9.94KK557 pKa = 10.56SSFRR561 pKa = 11.84LSDD564 pKa = 4.32LPTCVNGDD572 pKa = 4.0DD573 pKa = 3.69VLFWARR579 pKa = 11.84DD580 pKa = 3.43GAHH583 pKa = 5.54YY584 pKa = 9.02EE585 pKa = 3.33IWKK588 pKa = 10.19QITGEE593 pKa = 4.2CGLKK597 pKa = 10.43FSLGKK602 pKa = 10.35NYY604 pKa = 9.4TSRR607 pKa = 11.84HH608 pKa = 3.86VCVINSEE615 pKa = 4.34LYY617 pKa = 10.7RR618 pKa = 11.84FVKK621 pKa = 10.01DD622 pKa = 3.19TDD624 pKa = 4.1CQIHH628 pKa = 6.19RR629 pKa = 11.84PSPLFRR635 pKa = 11.84LEE637 pKa = 3.76KK638 pKa = 10.52ALNSRR643 pKa = 11.84LLCGGTRR650 pKa = 11.84SAASSGFDD658 pKa = 3.54PLMLSDD664 pKa = 4.7IDD666 pKa = 3.88LSIYY670 pKa = 10.42SSAVGGSAAFRR681 pKa = 11.84RR682 pKa = 11.84LVKK685 pKa = 10.48SHH687 pKa = 6.77LRR689 pKa = 11.84PHH691 pKa = 6.17QGVEE695 pKa = 3.84PSPKK699 pKa = 9.72KK700 pKa = 10.69VLAALRR706 pKa = 11.84KK707 pKa = 8.82RR708 pKa = 11.84YY709 pKa = 9.96RR710 pKa = 11.84NGGFSEE716 pKa = 6.23DD717 pKa = 3.44YY718 pKa = 10.83AKK720 pKa = 10.29WFNTIPSRR728 pKa = 11.84ALGLLEE734 pKa = 3.81QHH736 pKa = 6.91DD737 pKa = 4.74GEE739 pKa = 4.85IEE741 pKa = 3.82TDD743 pKa = 3.57LVVRR747 pKa = 11.84PKK749 pKa = 9.94MRR751 pKa = 11.84EE752 pKa = 3.55FMTKK756 pKa = 9.24VFNDD760 pKa = 3.51LQISKK765 pKa = 9.38LHH767 pKa = 5.94KK768 pKa = 9.69FARR771 pKa = 11.84ADD773 pKa = 3.35SRR775 pKa = 11.84VVKK778 pKa = 10.56NSPGFYY784 pKa = 10.09KK785 pKa = 10.15PQHH788 pKa = 6.17LGGLGLIPPINHH800 pKa = 5.41VFSIHH805 pKa = 6.51DD806 pKa = 3.7HH807 pKa = 6.96LEE809 pKa = 4.06IASLRR814 pKa = 11.84ALPAEE819 pKa = 3.95AHH821 pKa = 5.91QFSQGMTPKK830 pKa = 8.72MASVSFMEE838 pKa = 4.35SVRR841 pKa = 11.84AEE843 pKa = 3.68ILEE846 pKa = 4.23IQDD849 pKa = 3.32TLEE852 pKa = 3.94IEE854 pKa = 4.47RR855 pKa = 11.84ALLPAADD862 pKa = 3.72IEE864 pKa = 4.49SLRR867 pKa = 11.84FNGEE871 pKa = 3.63EE872 pKa = 3.99SQFWDD877 pKa = 3.2QEE879 pKa = 4.3FLVGFVDD886 pKa = 4.0PEE888 pKa = 4.17NVVVDD893 pKa = 3.69SEE895 pKa = 4.42SRR897 pKa = 11.84SRR899 pKa = 11.84KK900 pKa = 9.72EE901 pKa = 3.37SDD903 pKa = 3.15LNKK906 pKa = 10.16ILTSRR911 pKa = 11.84NWKK914 pKa = 9.25SRR916 pKa = 11.84RR917 pKa = 11.84LTGEE921 pKa = 3.64MRR923 pKa = 11.84RR924 pKa = 11.84QAALLEE930 pKa = 4.36KK931 pKa = 10.67ASGVKK936 pKa = 10.1CEE938 pKa = 4.27KK939 pKa = 10.17TFTDD943 pKa = 3.69ASHH946 pKa = 5.78GHH948 pKa = 5.85RR949 pKa = 11.84LRR951 pKa = 11.84YY952 pKa = 9.4LGAMPQGKK960 pKa = 8.05EE961 pKa = 3.94TVNLIEE967 pKa = 5.02GDD969 pKa = 2.82WHH971 pKa = 6.53LISRR975 pKa = 11.84RR976 pKa = 11.84PLPKK980 pKa = 10.12FHH982 pKa = 7.25
Molecular weight: 110.58 kDa
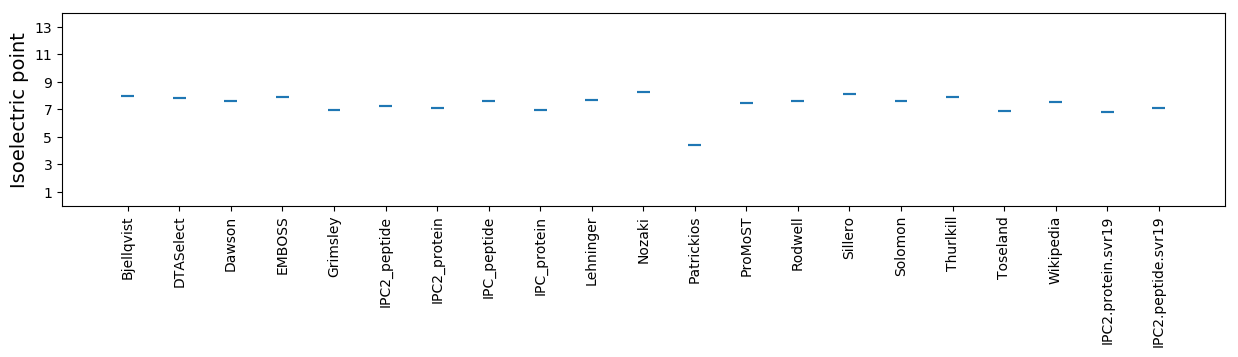
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI63|A0A1L3KI63_9VIRU RNA-dependent RNA polymerase OS=Changjiang narna-like virus 1 OX=1922776 PE=4 SV=1
MM1 pKa = 6.24MTEE4 pKa = 4.95SEE6 pKa = 4.69PLMCPDD12 pKa = 4.2SSSLSAPIGFAAALPKK28 pKa = 10.16KK29 pKa = 9.86AAAVLTIEE37 pKa = 5.42NIEE40 pKa = 3.74SWRR43 pKa = 11.84RR44 pKa = 11.84LTSASLPTAGAPSWHH59 pKa = 6.99PLVVVDD65 pKa = 4.38SQQEE69 pKa = 4.1ILSHH73 pKa = 6.81SEE75 pKa = 3.76ALGPPLDD82 pKa = 5.21LPLIDD87 pKa = 4.98CFIKK91 pKa = 10.4ASHH94 pKa = 6.48MIHH97 pKa = 7.04WYY99 pKa = 10.38NNTVEE104 pKa = 4.21IWAHH108 pKa = 4.88SLISGDD114 pKa = 3.66YY115 pKa = 11.09GEE117 pKa = 5.02SLVNKK122 pKa = 9.7GCILRR127 pKa = 11.84SCLSTPTCSRR137 pKa = 11.84MLGRR141 pKa = 11.84WTALAQFGKK150 pKa = 10.82LEE152 pKa = 4.7AYY154 pKa = 9.61LKK156 pKa = 10.1WVTANFFAEE165 pKa = 4.18SLLQKK170 pKa = 9.71EE171 pKa = 5.01LPPPPDD177 pKa = 3.15VFKK180 pKa = 11.33GILAKK185 pKa = 10.28IPGMKK190 pKa = 9.43RR191 pKa = 11.84PYY193 pKa = 9.69PGRR196 pKa = 11.84SWSSLFSRR204 pKa = 11.84THH206 pKa = 5.05SVKK209 pKa = 10.77SGLKK213 pKa = 8.72WRR215 pKa = 11.84FTFGKK220 pKa = 9.95DD221 pKa = 3.43FYY223 pKa = 8.44MTKK226 pKa = 10.2NASLPVEE233 pKa = 4.12EE234 pKa = 5.69SFVQANLEE242 pKa = 3.97KK243 pKa = 10.33HH244 pKa = 5.63QKK246 pKa = 8.96ILCGQIEE253 pKa = 4.67DD254 pKa = 4.29KK255 pKa = 11.06LSEE258 pKa = 3.87ATYY261 pKa = 11.12DD262 pKa = 3.87EE263 pKa = 4.44VCEE266 pKa = 4.88AIQLCADD273 pKa = 4.5EE274 pKa = 5.6IFGKK278 pKa = 9.85PYY280 pKa = 9.97EE281 pKa = 4.33VPSKK285 pKa = 10.6FISSDD290 pKa = 3.62LEE292 pKa = 3.57ITEE295 pKa = 4.61NIRR298 pKa = 11.84SFSRR302 pKa = 11.84PEE304 pKa = 3.57IKK306 pKa = 10.35CPSRR310 pKa = 11.84LPSFGASFHH319 pKa = 6.34GNRR322 pKa = 11.84GDD324 pKa = 4.37GGACGDD330 pKa = 4.29LLRR333 pKa = 11.84NYY335 pKa = 10.01HH336 pKa = 7.26DD337 pKa = 4.05IGSLPEE343 pKa = 4.59PNDD346 pKa = 3.95GYY348 pKa = 11.46LWGYY352 pKa = 7.56AQKK355 pKa = 10.73NGSEE359 pKa = 4.12LCEE362 pKa = 3.87VRR364 pKa = 11.84TLHH367 pKa = 7.27DD368 pKa = 3.82PEE370 pKa = 5.26LFFEE374 pKa = 5.83AEE376 pKa = 4.37KK377 pKa = 10.33QWSRR381 pKa = 11.84EE382 pKa = 3.84ALFLSSEE389 pKa = 4.28GVNAHH394 pKa = 5.43VVPLIEE400 pKa = 4.24PFKK403 pKa = 11.16VRR405 pKa = 11.84TITKK409 pKa = 8.21GQAEE413 pKa = 4.76IYY415 pKa = 9.77HH416 pKa = 6.99LARR419 pKa = 11.84RR420 pKa = 11.84WQKK423 pKa = 10.27IIHH426 pKa = 5.65SRR428 pKa = 11.84MRR430 pKa = 11.84QHH432 pKa = 7.2PNFKK436 pKa = 10.75LIGQPCNGAFISQIFGNSKK455 pKa = 9.95LFNYY459 pKa = 10.44YY460 pKa = 10.24GDD462 pKa = 3.6KK463 pKa = 11.21NGFFVSGDD471 pKa = 3.67YY472 pKa = 11.0EE473 pKa = 4.48SATDD477 pKa = 4.34LLNPALSEE485 pKa = 4.03FAQEE489 pKa = 4.06QISIRR494 pKa = 11.84LGIPLEE500 pKa = 4.15DD501 pKa = 3.32QYY503 pKa = 12.06VLKK506 pKa = 10.76QCLTGHH512 pKa = 6.49RR513 pKa = 11.84LRR515 pKa = 11.84YY516 pKa = 9.15EE517 pKa = 3.85KK518 pKa = 10.54EE519 pKa = 3.95GQLFEE524 pKa = 4.32QTWGQLMGSPTSFPVLCLVNMAATLLSYY552 pKa = 10.85NRR554 pKa = 11.84AYY556 pKa = 9.94KK557 pKa = 10.56SSFRR561 pKa = 11.84LSDD564 pKa = 4.32LPTCVNGDD572 pKa = 4.0DD573 pKa = 3.69VLFWARR579 pKa = 11.84DD580 pKa = 3.43GAHH583 pKa = 5.54YY584 pKa = 9.02EE585 pKa = 3.33IWKK588 pKa = 10.19QITGEE593 pKa = 4.2CGLKK597 pKa = 10.43FSLGKK602 pKa = 10.35NYY604 pKa = 9.4TSRR607 pKa = 11.84HH608 pKa = 3.86VCVINSEE615 pKa = 4.34LYY617 pKa = 10.7RR618 pKa = 11.84FVKK621 pKa = 10.01DD622 pKa = 3.19TDD624 pKa = 4.1CQIHH628 pKa = 6.19RR629 pKa = 11.84PSPLFRR635 pKa = 11.84LEE637 pKa = 3.76KK638 pKa = 10.52ALNSRR643 pKa = 11.84LLCGGTRR650 pKa = 11.84SAASSGFDD658 pKa = 3.54PLMLSDD664 pKa = 4.7IDD666 pKa = 3.88LSIYY670 pKa = 10.42SSAVGGSAAFRR681 pKa = 11.84RR682 pKa = 11.84LVKK685 pKa = 10.48SHH687 pKa = 6.77LRR689 pKa = 11.84PHH691 pKa = 6.17QGVEE695 pKa = 3.84PSPKK699 pKa = 9.72KK700 pKa = 10.69VLAALRR706 pKa = 11.84KK707 pKa = 8.82RR708 pKa = 11.84YY709 pKa = 9.96RR710 pKa = 11.84NGGFSEE716 pKa = 6.23DD717 pKa = 3.44YY718 pKa = 10.83AKK720 pKa = 10.29WFNTIPSRR728 pKa = 11.84ALGLLEE734 pKa = 3.81QHH736 pKa = 6.91DD737 pKa = 4.74GEE739 pKa = 4.85IEE741 pKa = 3.82TDD743 pKa = 3.57LVVRR747 pKa = 11.84PKK749 pKa = 9.94MRR751 pKa = 11.84EE752 pKa = 3.55FMTKK756 pKa = 9.24VFNDD760 pKa = 3.51LQISKK765 pKa = 9.38LHH767 pKa = 5.94KK768 pKa = 9.69FARR771 pKa = 11.84ADD773 pKa = 3.35SRR775 pKa = 11.84VVKK778 pKa = 10.56NSPGFYY784 pKa = 10.09KK785 pKa = 10.15PQHH788 pKa = 6.17LGGLGLIPPINHH800 pKa = 5.41VFSIHH805 pKa = 6.51DD806 pKa = 3.7HH807 pKa = 6.96LEE809 pKa = 4.06IASLRR814 pKa = 11.84ALPAEE819 pKa = 3.95AHH821 pKa = 5.91QFSQGMTPKK830 pKa = 8.72MASVSFMEE838 pKa = 4.35SVRR841 pKa = 11.84AEE843 pKa = 3.68ILEE846 pKa = 4.23IQDD849 pKa = 3.32TLEE852 pKa = 3.94IEE854 pKa = 4.47RR855 pKa = 11.84ALLPAADD862 pKa = 3.72IEE864 pKa = 4.49SLRR867 pKa = 11.84FNGEE871 pKa = 3.63EE872 pKa = 3.99SQFWDD877 pKa = 3.2QEE879 pKa = 4.3FLVGFVDD886 pKa = 4.0PEE888 pKa = 4.17NVVVDD893 pKa = 3.69SEE895 pKa = 4.42SRR897 pKa = 11.84SRR899 pKa = 11.84KK900 pKa = 9.72EE901 pKa = 3.37SDD903 pKa = 3.15LNKK906 pKa = 10.16ILTSRR911 pKa = 11.84NWKK914 pKa = 9.25SRR916 pKa = 11.84RR917 pKa = 11.84LTGEE921 pKa = 3.64MRR923 pKa = 11.84RR924 pKa = 11.84QAALLEE930 pKa = 4.36KK931 pKa = 10.67ASGVKK936 pKa = 10.1CEE938 pKa = 4.27KK939 pKa = 10.17TFTDD943 pKa = 3.69ASHH946 pKa = 5.78GHH948 pKa = 5.85RR949 pKa = 11.84LRR951 pKa = 11.84YY952 pKa = 9.4LGAMPQGKK960 pKa = 8.05EE961 pKa = 3.94TVNLIEE967 pKa = 5.02GDD969 pKa = 2.82WHH971 pKa = 6.53LISRR975 pKa = 11.84RR976 pKa = 11.84PLPKK980 pKa = 10.12FHH982 pKa = 7.25
MM1 pKa = 6.24MTEE4 pKa = 4.95SEE6 pKa = 4.69PLMCPDD12 pKa = 4.2SSSLSAPIGFAAALPKK28 pKa = 10.16KK29 pKa = 9.86AAAVLTIEE37 pKa = 5.42NIEE40 pKa = 3.74SWRR43 pKa = 11.84RR44 pKa = 11.84LTSASLPTAGAPSWHH59 pKa = 6.99PLVVVDD65 pKa = 4.38SQQEE69 pKa = 4.1ILSHH73 pKa = 6.81SEE75 pKa = 3.76ALGPPLDD82 pKa = 5.21LPLIDD87 pKa = 4.98CFIKK91 pKa = 10.4ASHH94 pKa = 6.48MIHH97 pKa = 7.04WYY99 pKa = 10.38NNTVEE104 pKa = 4.21IWAHH108 pKa = 4.88SLISGDD114 pKa = 3.66YY115 pKa = 11.09GEE117 pKa = 5.02SLVNKK122 pKa = 9.7GCILRR127 pKa = 11.84SCLSTPTCSRR137 pKa = 11.84MLGRR141 pKa = 11.84WTALAQFGKK150 pKa = 10.82LEE152 pKa = 4.7AYY154 pKa = 9.61LKK156 pKa = 10.1WVTANFFAEE165 pKa = 4.18SLLQKK170 pKa = 9.71EE171 pKa = 5.01LPPPPDD177 pKa = 3.15VFKK180 pKa = 11.33GILAKK185 pKa = 10.28IPGMKK190 pKa = 9.43RR191 pKa = 11.84PYY193 pKa = 9.69PGRR196 pKa = 11.84SWSSLFSRR204 pKa = 11.84THH206 pKa = 5.05SVKK209 pKa = 10.77SGLKK213 pKa = 8.72WRR215 pKa = 11.84FTFGKK220 pKa = 9.95DD221 pKa = 3.43FYY223 pKa = 8.44MTKK226 pKa = 10.2NASLPVEE233 pKa = 4.12EE234 pKa = 5.69SFVQANLEE242 pKa = 3.97KK243 pKa = 10.33HH244 pKa = 5.63QKK246 pKa = 8.96ILCGQIEE253 pKa = 4.67DD254 pKa = 4.29KK255 pKa = 11.06LSEE258 pKa = 3.87ATYY261 pKa = 11.12DD262 pKa = 3.87EE263 pKa = 4.44VCEE266 pKa = 4.88AIQLCADD273 pKa = 4.5EE274 pKa = 5.6IFGKK278 pKa = 9.85PYY280 pKa = 9.97EE281 pKa = 4.33VPSKK285 pKa = 10.6FISSDD290 pKa = 3.62LEE292 pKa = 3.57ITEE295 pKa = 4.61NIRR298 pKa = 11.84SFSRR302 pKa = 11.84PEE304 pKa = 3.57IKK306 pKa = 10.35CPSRR310 pKa = 11.84LPSFGASFHH319 pKa = 6.34GNRR322 pKa = 11.84GDD324 pKa = 4.37GGACGDD330 pKa = 4.29LLRR333 pKa = 11.84NYY335 pKa = 10.01HH336 pKa = 7.26DD337 pKa = 4.05IGSLPEE343 pKa = 4.59PNDD346 pKa = 3.95GYY348 pKa = 11.46LWGYY352 pKa = 7.56AQKK355 pKa = 10.73NGSEE359 pKa = 4.12LCEE362 pKa = 3.87VRR364 pKa = 11.84TLHH367 pKa = 7.27DD368 pKa = 3.82PEE370 pKa = 5.26LFFEE374 pKa = 5.83AEE376 pKa = 4.37KK377 pKa = 10.33QWSRR381 pKa = 11.84EE382 pKa = 3.84ALFLSSEE389 pKa = 4.28GVNAHH394 pKa = 5.43VVPLIEE400 pKa = 4.24PFKK403 pKa = 11.16VRR405 pKa = 11.84TITKK409 pKa = 8.21GQAEE413 pKa = 4.76IYY415 pKa = 9.77HH416 pKa = 6.99LARR419 pKa = 11.84RR420 pKa = 11.84WQKK423 pKa = 10.27IIHH426 pKa = 5.65SRR428 pKa = 11.84MRR430 pKa = 11.84QHH432 pKa = 7.2PNFKK436 pKa = 10.75LIGQPCNGAFISQIFGNSKK455 pKa = 9.95LFNYY459 pKa = 10.44YY460 pKa = 10.24GDD462 pKa = 3.6KK463 pKa = 11.21NGFFVSGDD471 pKa = 3.67YY472 pKa = 11.0EE473 pKa = 4.48SATDD477 pKa = 4.34LLNPALSEE485 pKa = 4.03FAQEE489 pKa = 4.06QISIRR494 pKa = 11.84LGIPLEE500 pKa = 4.15DD501 pKa = 3.32QYY503 pKa = 12.06VLKK506 pKa = 10.76QCLTGHH512 pKa = 6.49RR513 pKa = 11.84LRR515 pKa = 11.84YY516 pKa = 9.15EE517 pKa = 3.85KK518 pKa = 10.54EE519 pKa = 3.95GQLFEE524 pKa = 4.32QTWGQLMGSPTSFPVLCLVNMAATLLSYY552 pKa = 10.85NRR554 pKa = 11.84AYY556 pKa = 9.94KK557 pKa = 10.56SSFRR561 pKa = 11.84LSDD564 pKa = 4.32LPTCVNGDD572 pKa = 4.0DD573 pKa = 3.69VLFWARR579 pKa = 11.84DD580 pKa = 3.43GAHH583 pKa = 5.54YY584 pKa = 9.02EE585 pKa = 3.33IWKK588 pKa = 10.19QITGEE593 pKa = 4.2CGLKK597 pKa = 10.43FSLGKK602 pKa = 10.35NYY604 pKa = 9.4TSRR607 pKa = 11.84HH608 pKa = 3.86VCVINSEE615 pKa = 4.34LYY617 pKa = 10.7RR618 pKa = 11.84FVKK621 pKa = 10.01DD622 pKa = 3.19TDD624 pKa = 4.1CQIHH628 pKa = 6.19RR629 pKa = 11.84PSPLFRR635 pKa = 11.84LEE637 pKa = 3.76KK638 pKa = 10.52ALNSRR643 pKa = 11.84LLCGGTRR650 pKa = 11.84SAASSGFDD658 pKa = 3.54PLMLSDD664 pKa = 4.7IDD666 pKa = 3.88LSIYY670 pKa = 10.42SSAVGGSAAFRR681 pKa = 11.84RR682 pKa = 11.84LVKK685 pKa = 10.48SHH687 pKa = 6.77LRR689 pKa = 11.84PHH691 pKa = 6.17QGVEE695 pKa = 3.84PSPKK699 pKa = 9.72KK700 pKa = 10.69VLAALRR706 pKa = 11.84KK707 pKa = 8.82RR708 pKa = 11.84YY709 pKa = 9.96RR710 pKa = 11.84NGGFSEE716 pKa = 6.23DD717 pKa = 3.44YY718 pKa = 10.83AKK720 pKa = 10.29WFNTIPSRR728 pKa = 11.84ALGLLEE734 pKa = 3.81QHH736 pKa = 6.91DD737 pKa = 4.74GEE739 pKa = 4.85IEE741 pKa = 3.82TDD743 pKa = 3.57LVVRR747 pKa = 11.84PKK749 pKa = 9.94MRR751 pKa = 11.84EE752 pKa = 3.55FMTKK756 pKa = 9.24VFNDD760 pKa = 3.51LQISKK765 pKa = 9.38LHH767 pKa = 5.94KK768 pKa = 9.69FARR771 pKa = 11.84ADD773 pKa = 3.35SRR775 pKa = 11.84VVKK778 pKa = 10.56NSPGFYY784 pKa = 10.09KK785 pKa = 10.15PQHH788 pKa = 6.17LGGLGLIPPINHH800 pKa = 5.41VFSIHH805 pKa = 6.51DD806 pKa = 3.7HH807 pKa = 6.96LEE809 pKa = 4.06IASLRR814 pKa = 11.84ALPAEE819 pKa = 3.95AHH821 pKa = 5.91QFSQGMTPKK830 pKa = 8.72MASVSFMEE838 pKa = 4.35SVRR841 pKa = 11.84AEE843 pKa = 3.68ILEE846 pKa = 4.23IQDD849 pKa = 3.32TLEE852 pKa = 3.94IEE854 pKa = 4.47RR855 pKa = 11.84ALLPAADD862 pKa = 3.72IEE864 pKa = 4.49SLRR867 pKa = 11.84FNGEE871 pKa = 3.63EE872 pKa = 3.99SQFWDD877 pKa = 3.2QEE879 pKa = 4.3FLVGFVDD886 pKa = 4.0PEE888 pKa = 4.17NVVVDD893 pKa = 3.69SEE895 pKa = 4.42SRR897 pKa = 11.84SRR899 pKa = 11.84KK900 pKa = 9.72EE901 pKa = 3.37SDD903 pKa = 3.15LNKK906 pKa = 10.16ILTSRR911 pKa = 11.84NWKK914 pKa = 9.25SRR916 pKa = 11.84RR917 pKa = 11.84LTGEE921 pKa = 3.64MRR923 pKa = 11.84RR924 pKa = 11.84QAALLEE930 pKa = 4.36KK931 pKa = 10.67ASGVKK936 pKa = 10.1CEE938 pKa = 4.27KK939 pKa = 10.17TFTDD943 pKa = 3.69ASHH946 pKa = 5.78GHH948 pKa = 5.85RR949 pKa = 11.84LRR951 pKa = 11.84YY952 pKa = 9.4LGAMPQGKK960 pKa = 8.05EE961 pKa = 3.94TVNLIEE967 pKa = 5.02GDD969 pKa = 2.82WHH971 pKa = 6.53LISRR975 pKa = 11.84RR976 pKa = 11.84PLPKK980 pKa = 10.12FHH982 pKa = 7.25
Molecular weight: 110.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
982 |
982 |
982 |
982.0 |
110.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.823 ± 0.0 | 2.037 ± 0.0 |
4.379 ± 0.0 | 7.026 ± 0.0 |
5.193 ± 0.0 | 6.721 ± 0.0 |
3.157 ± 0.0 | 5.295 ± 0.0 |
5.703 ± 0.0 | 10.794 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.833 ± 0.0 | 3.564 ± 0.0 |
5.601 ± 0.0 | 3.564 ± 0.0 |
6.008 ± 0.0 | 9.369 ± 0.0 |
3.768 ± 0.0 | 4.684 ± 0.0 |
1.833 ± 0.0 | 2.648 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
