
Hyaloperonospora arabidopsidis (strain Emoy2) (Downy mildew agent) (Peronospora arabidopsidis)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Peronosporales; Peronosporaceae; Hyaloperonospora; Hyaloperonospora arabidopsidis
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
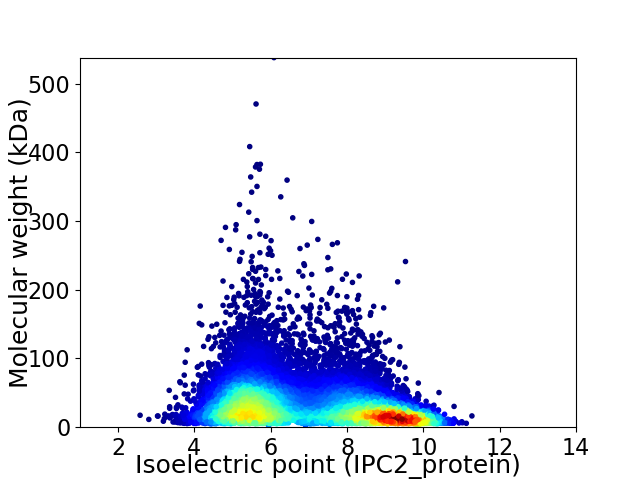
Virtual 2D-PAGE plot for 13837 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
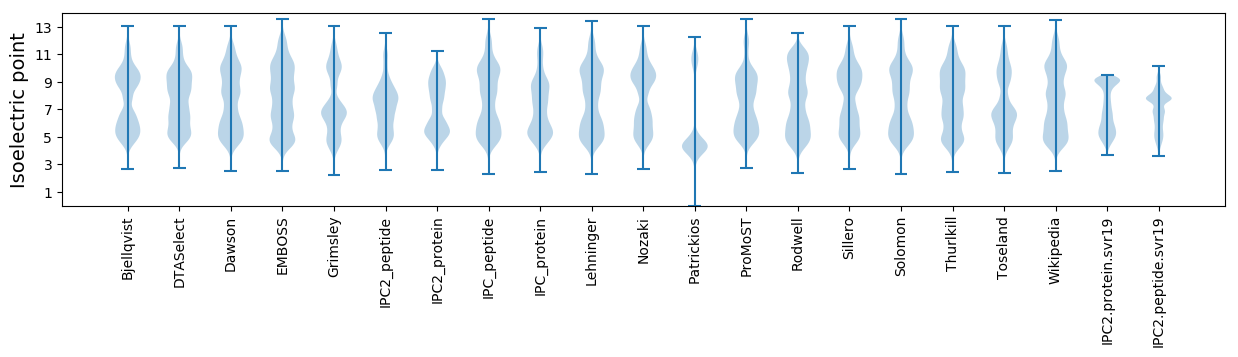
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M4C067|M4C067_HYAAE Uncharacterized protein OS=Hyaloperonospora arabidopsidis (strain Emoy2) OX=559515 PE=4 SV=1
MM1 pKa = 7.48TISKK5 pKa = 7.62THH7 pKa = 5.67HH8 pKa = 6.09TLLLGAAALALDD20 pKa = 3.91AVNAHH25 pKa = 7.17GYY27 pKa = 9.34CSRR30 pKa = 11.84PAVTFPNGGDD40 pKa = 3.27TTQFVATIEE49 pKa = 4.37SSASGLPGVFTGSPADD65 pKa = 3.43NAAAFWTAFGSSKK78 pKa = 10.24FSSIKK83 pKa = 10.68DD84 pKa = 3.41LATSLGQIVAKK95 pKa = 10.34GATLEE100 pKa = 4.98CGLTDD105 pKa = 4.85PNGTPQPIPDD115 pKa = 3.78EE116 pKa = 4.89LEE118 pKa = 3.92WSHH121 pKa = 7.24SGTEE125 pKa = 4.27GFTASHH131 pKa = 7.03EE132 pKa = 4.82GPCEE136 pKa = 3.74AWCDD140 pKa = 3.75DD141 pKa = 3.62TQVFQDD147 pKa = 4.25EE148 pKa = 4.15NCAAHH153 pKa = 6.37FTTAPAKK160 pKa = 9.85MPYY163 pKa = 10.38DD164 pKa = 3.47KK165 pKa = 10.72AKK167 pKa = 10.72CSGASRR173 pKa = 11.84FTFYY177 pKa = 10.5WLALHH182 pKa = 6.73SSTWQVYY189 pKa = 6.79VNCAALEE196 pKa = 4.45GGSGDD201 pKa = 3.82MYY203 pKa = 10.37STSQTEE209 pKa = 4.33VNASEE214 pKa = 4.44STEE217 pKa = 3.92QSSPPTSSSDD227 pKa = 3.16TPTAVVAPDD236 pKa = 3.14QSYY239 pKa = 11.38SFTGTTAAPSTTDD252 pKa = 3.25VPVNTSKK259 pKa = 9.69STGEE263 pKa = 4.14STTPAPSNATAPADD277 pKa = 3.6VVPTSTPSPTGEE289 pKa = 3.97PSFTFADD296 pKa = 4.0TNAMPNTTEE305 pKa = 3.96AVAPNTTKK313 pKa = 10.54IIGSNTSEE321 pKa = 4.18TVAPSTPAPTDD332 pKa = 3.53PSNTPSAPIATPTTEE347 pKa = 4.34VISPTGDD354 pKa = 2.44ITTSSNSDD362 pKa = 3.39VNSVGVVGIKK372 pKa = 10.09DD373 pKa = 3.58DD374 pKa = 4.58CGSLDD379 pKa = 3.52MAGEE383 pKa = 4.26GDD385 pKa = 3.85NVVKK389 pKa = 10.91ANTDD393 pKa = 2.84ISSSSNSGVGPDD405 pKa = 3.44VNTVGVVEE413 pKa = 4.66TTATPNTTEE422 pKa = 4.95AIAPNTTGAIAPNTSKK438 pKa = 10.73IIGSNTSEE446 pKa = 4.18TVAPSTPAPTDD457 pKa = 3.5PSNTPTATTTTPTTEE472 pKa = 4.34VISPTGGITTSSNSGVGSDD491 pKa = 3.7VNAVSVVGIEE501 pKa = 4.34DD502 pKa = 3.79DD503 pKa = 4.55CGSLDD508 pKa = 3.71MAGEE512 pKa = 4.18GDD514 pKa = 3.6NRR516 pKa = 11.84VEE518 pKa = 4.21TSTVPVPDD526 pKa = 4.17EE527 pKa = 4.59DD528 pKa = 4.58CGSFDD533 pKa = 3.46IAGSEE538 pKa = 4.09DD539 pKa = 3.71MEE541 pKa = 5.11DD542 pKa = 4.27CGSLDD547 pKa = 3.19IAGGSLDD554 pKa = 3.72GVGSSGIDD562 pKa = 3.23NRR564 pKa = 11.84ITSDD568 pKa = 3.39SVGSFAGEE576 pKa = 3.79VNPAISEE583 pKa = 4.19SSLNFDD589 pKa = 4.53DD590 pKa = 4.54VDD592 pKa = 3.5RR593 pKa = 11.84GYY595 pKa = 9.53TGGKK599 pKa = 8.18VQAPYY604 pKa = 10.64QYY606 pKa = 11.71
MM1 pKa = 7.48TISKK5 pKa = 7.62THH7 pKa = 5.67HH8 pKa = 6.09TLLLGAAALALDD20 pKa = 3.91AVNAHH25 pKa = 7.17GYY27 pKa = 9.34CSRR30 pKa = 11.84PAVTFPNGGDD40 pKa = 3.27TTQFVATIEE49 pKa = 4.37SSASGLPGVFTGSPADD65 pKa = 3.43NAAAFWTAFGSSKK78 pKa = 10.24FSSIKK83 pKa = 10.68DD84 pKa = 3.41LATSLGQIVAKK95 pKa = 10.34GATLEE100 pKa = 4.98CGLTDD105 pKa = 4.85PNGTPQPIPDD115 pKa = 3.78EE116 pKa = 4.89LEE118 pKa = 3.92WSHH121 pKa = 7.24SGTEE125 pKa = 4.27GFTASHH131 pKa = 7.03EE132 pKa = 4.82GPCEE136 pKa = 3.74AWCDD140 pKa = 3.75DD141 pKa = 3.62TQVFQDD147 pKa = 4.25EE148 pKa = 4.15NCAAHH153 pKa = 6.37FTTAPAKK160 pKa = 9.85MPYY163 pKa = 10.38DD164 pKa = 3.47KK165 pKa = 10.72AKK167 pKa = 10.72CSGASRR173 pKa = 11.84FTFYY177 pKa = 10.5WLALHH182 pKa = 6.73SSTWQVYY189 pKa = 6.79VNCAALEE196 pKa = 4.45GGSGDD201 pKa = 3.82MYY203 pKa = 10.37STSQTEE209 pKa = 4.33VNASEE214 pKa = 4.44STEE217 pKa = 3.92QSSPPTSSSDD227 pKa = 3.16TPTAVVAPDD236 pKa = 3.14QSYY239 pKa = 11.38SFTGTTAAPSTTDD252 pKa = 3.25VPVNTSKK259 pKa = 9.69STGEE263 pKa = 4.14STTPAPSNATAPADD277 pKa = 3.6VVPTSTPSPTGEE289 pKa = 3.97PSFTFADD296 pKa = 4.0TNAMPNTTEE305 pKa = 3.96AVAPNTTKK313 pKa = 10.54IIGSNTSEE321 pKa = 4.18TVAPSTPAPTDD332 pKa = 3.53PSNTPSAPIATPTTEE347 pKa = 4.34VISPTGDD354 pKa = 2.44ITTSSNSDD362 pKa = 3.39VNSVGVVGIKK372 pKa = 10.09DD373 pKa = 3.58DD374 pKa = 4.58CGSLDD379 pKa = 3.52MAGEE383 pKa = 4.26GDD385 pKa = 3.85NVVKK389 pKa = 10.91ANTDD393 pKa = 2.84ISSSSNSGVGPDD405 pKa = 3.44VNTVGVVEE413 pKa = 4.66TTATPNTTEE422 pKa = 4.95AIAPNTTGAIAPNTSKK438 pKa = 10.73IIGSNTSEE446 pKa = 4.18TVAPSTPAPTDD457 pKa = 3.5PSNTPTATTTTPTTEE472 pKa = 4.34VISPTGGITTSSNSGVGSDD491 pKa = 3.7VNAVSVVGIEE501 pKa = 4.34DD502 pKa = 3.79DD503 pKa = 4.55CGSLDD508 pKa = 3.71MAGEE512 pKa = 4.18GDD514 pKa = 3.6NRR516 pKa = 11.84VEE518 pKa = 4.21TSTVPVPDD526 pKa = 4.17EE527 pKa = 4.59DD528 pKa = 4.58CGSFDD533 pKa = 3.46IAGSEE538 pKa = 4.09DD539 pKa = 3.71MEE541 pKa = 5.11DD542 pKa = 4.27CGSLDD547 pKa = 3.19IAGGSLDD554 pKa = 3.72GVGSSGIDD562 pKa = 3.23NRR564 pKa = 11.84ITSDD568 pKa = 3.39SVGSFAGEE576 pKa = 3.79VNPAISEE583 pKa = 4.19SSLNFDD589 pKa = 4.53DD590 pKa = 4.54VDD592 pKa = 3.5RR593 pKa = 11.84GYY595 pKa = 9.53TGGKK599 pKa = 8.18VQAPYY604 pKa = 10.64QYY606 pKa = 11.71
Molecular weight: 61.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M4B3H1|M4B3H1_HYAAE Uncharacterized protein OS=Hyaloperonospora arabidopsidis (strain Emoy2) OX=559515 PE=4 SV=1
MM1 pKa = 7.16VRR3 pKa = 11.84RR4 pKa = 11.84LRR6 pKa = 11.84SGAVPARR13 pKa = 11.84LGRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84LIARR22 pKa = 11.84VWTKK26 pKa = 10.59RR27 pKa = 11.84DD28 pKa = 3.1QTTRR32 pKa = 11.84RR33 pKa = 11.84HH34 pKa = 4.41QRR36 pKa = 11.84RR37 pKa = 11.84PRR39 pKa = 11.84AIAAAMALTRR49 pKa = 11.84HH50 pKa = 5.98KK51 pKa = 10.12LAKK54 pKa = 9.44GVRR57 pKa = 11.84GVRR60 pKa = 11.84ILLPAVEE67 pKa = 4.48TPRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84LLRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84IPRR81 pKa = 11.84HH82 pKa = 3.73HH83 pKa = 6.02HH84 pKa = 3.57QRR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84VTLRR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84LWPLRR104 pKa = 11.84SRR106 pKa = 11.84RR107 pKa = 11.84LRR109 pKa = 11.84ARR111 pKa = 11.84TVHH114 pKa = 6.82RR115 pKa = 11.84PLQRR119 pKa = 11.84VRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84GRR125 pKa = 11.84LQTCRR130 pKa = 11.84RR131 pKa = 11.84LL132 pKa = 3.43
MM1 pKa = 7.16VRR3 pKa = 11.84RR4 pKa = 11.84LRR6 pKa = 11.84SGAVPARR13 pKa = 11.84LGRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84LIARR22 pKa = 11.84VWTKK26 pKa = 10.59RR27 pKa = 11.84DD28 pKa = 3.1QTTRR32 pKa = 11.84RR33 pKa = 11.84HH34 pKa = 4.41QRR36 pKa = 11.84RR37 pKa = 11.84PRR39 pKa = 11.84AIAAAMALTRR49 pKa = 11.84HH50 pKa = 5.98KK51 pKa = 10.12LAKK54 pKa = 9.44GVRR57 pKa = 11.84GVRR60 pKa = 11.84ILLPAVEE67 pKa = 4.48TPRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84LLRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84IPRR81 pKa = 11.84HH82 pKa = 3.73HH83 pKa = 6.02HH84 pKa = 3.57QRR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84VTLRR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84LWPLRR104 pKa = 11.84SRR106 pKa = 11.84RR107 pKa = 11.84LRR109 pKa = 11.84ARR111 pKa = 11.84TVHH114 pKa = 6.82RR115 pKa = 11.84PLQRR119 pKa = 11.84VRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84GRR125 pKa = 11.84LQTCRR130 pKa = 11.84RR131 pKa = 11.84LL132 pKa = 3.43
Molecular weight: 16.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4494721 |
49 |
4812 |
324.8 |
36.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.569 ± 0.021 | 1.894 ± 0.011 |
5.672 ± 0.015 | 6.331 ± 0.023 |
3.618 ± 0.015 | 6.061 ± 0.018 |
2.561 ± 0.011 | 4.081 ± 0.015 |
5.083 ± 0.022 | 9.452 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.563 ± 0.009 | 3.331 ± 0.011 |
4.453 ± 0.017 | 4.143 ± 0.016 |
6.697 ± 0.023 | 8.416 ± 0.025 |
5.904 ± 0.015 | 7.406 ± 0.018 |
1.211 ± 0.007 | 2.529 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
